| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,686,410 – 1,686,514 |
| Length | 104 |
| Max. P | 0.911786 |

| Location | 1,686,410 – 1,686,514 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
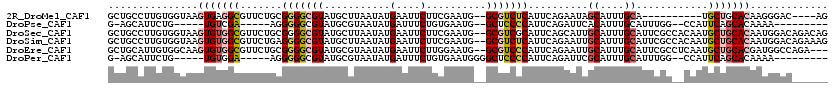
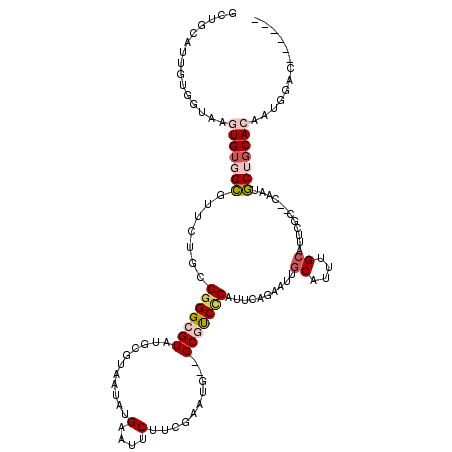
>2R_DroMel_CAF1 1686410 104 - 20766785 GCUGCCUUGUGGUAAGUGAGGCGUUCUGCGGGGCGUAUGCUUAAUAUGAAUUCUUCGAAUG--GCGUCUCAUUCAGAAUAGCAUUUGCA----------UGCUGCACAAGGGAC----AG .((((((((((...(((((((((((...((((((((((.....)))))....)))))...)--)))))))))).....((((((....)----------))))))))))))..)----)) ( -35.90) >DroPse_CAF1 102682 96 - 1 G-AGCAUUCUG-----UGUCGA-----AGGGGGCGUAUGCGUAAUAUGAUUUCUGUGAAUG--GCUCCCCAUUCAGAUUCACAUUUGCAUUUGG--CCAUUCAGCACAAAA--------- .-.......((-----(((.((-----(..((.((.(((((.(((.(((..(((..(((((--(....))))))))).))).)))))))).)).--)).))).)))))...--------- ( -29.10) >DroSec_CAF1 71651 118 - 1 GCUGCCUUGUGGUAAGUGUGGCGUUCUGCGGGGCGUAUGCUUAAUAUGAAUUCUUCGAAUG--GCGUCGCAUUCAGCAUUGCAUUUGCAUUCGCCACAAUGCUGCACAAUGGACAGACAG .(((((.((..(((..(((((((...(((((((((.(((((......((.....))(((((--......))))))))))))).))))))..))))))).)))..))....)).))).... ( -41.00) >DroSim_CAF1 69103 118 - 1 GCUGCCUUGUGGUAAGUGUGGCGUUCUGAGGGGCGUAUGCUUAAUAUGAAUUCUUCGAAUG--GCGUCUCAUUCAGAAUUGCAUUUGCAUUCGCCACAAUGCUGCACAAUGGACAGAAAG .(((((.((..(((..((((((((((((((((.(((((.....)))))...)))))(((((--(....)))))))))).(((....)))..))))))).)))..))....)).))).... ( -38.80) >DroEre_CAF1 76960 115 - 1 GCUGCAUUGUGGCAAGUGUGGCGUUCUGCGGGGCGUAUGCGUAAUAUGAAUUCUUGGAAUG--GCGUCCCAUUCAGAAUUGCAUUUGCAUUCGCCUCAAUGCUGCACGAUGGCCAGA--- .........((((..(((..(((((....((((((.(((((....(((((((((..(((((--(....)))))))))))).))).))))).)))))))))))..)))....))))..--- ( -47.30) >DroPer_CAF1 96554 98 - 1 G-AGCAUUCUG-----UGUGGA-----AGGGGGCGUAUGCGUAAUAUGAUUUCUGUGAAUGGGGCUCCCCAUUCAGAUUCGCAUUUGCAUUUGG--CCAUUCAGCACAAAA--------- .-.......((-----(((.((-----(..((.((.(((((.(((.(((..(((..(((((((....)))))))))).))).)))))))).)).--)).))).)))))...--------- ( -33.90) >consensus GCUGCAUUGUGGUAAGUGUGGCGUUCUGCGGGGCGUAUGCGUAAUAUGAAUUCUUCGAAUG__GCGUCCCAUUCAGAAUUGCAUUUGCAUUCGC__CAAUGCUGCACAAUGGAC______ ...............(((((((.......(((((((...........(....)..........)))))))..........((....))............)))))))............. (-15.00 = -15.67 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:28 2006