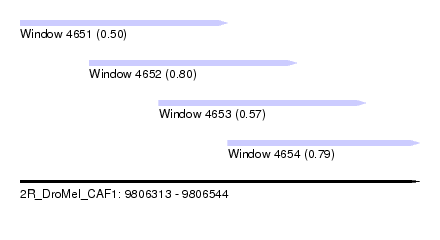
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,806,313 – 9,806,544 |
| Length | 231 |
| Max. P | 0.801027 |

| Location | 9,806,313 – 9,806,433 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -32.59 |
| Consensus MFE | -28.99 |
| Energy contribution | -29.27 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9806313 120 + 20766785 GUCGUUGUACGGCACUCCGAAAGAGGAGCCCAUGCCCAACAUACCGAUCGUGUCGGAAAAAGUCUCUGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACGGACAACCG ...(((((.((((....(....).((((((.(((.........(((((...)))))..........(((.((((..((....))..)))))))..))))))))).))))....))))).. ( -32.50) >DroVir_CAF1 48020 120 + 1 GUCGUUGUACGGCACUCCGAAAGAGGAGCCAAUGCCCAAUAUACCGAUAGUCUCGGAAAAAGUCUCGGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCG (((..(((.((((....(....).(((((((............((((.....))))...........((.((((..((....))..)))))).....))))))).))))))))))..... ( -32.20) >DroPse_CAF1 42898 120 + 1 GUCGUUGUACGGCACUCCGAAAGAGGAGCCGAUGCCGAACAUACCGAUCGUGUCGGAAAAAGUCUCUGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACGGACAAUCG (((..(((.((((....(....).(((((((((.((((.(((.......))))))).....)))..(((.((((..((....))..))))))).....)))))).))))))))))..... ( -34.10) >DroGri_CAF1 53063 120 + 1 GUCGUUGUACGGCACUCCGAAAGAGGAGCCAAUGCCCAACAUACCAAUAGUCUCGGAAAAAGUCUCGGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCG (((..(((.((((....(....).((((((((((.....)))..((((.(.((.(((((..((....))...))))).....)).).))))......))))))).))))))))))..... ( -30.60) >DroWil_CAF1 53166 120 + 1 GUCGUUGUACGGCACUCCGAAAGAGGAGCCCAUGCCCAAUAUACCUAUCGUGUCCGAGAAGGUCUCUGCUAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCG (((..(((.((((....(....).((((((.(((........((((.(((....)))..))))...(((.((((..((....))..)))))))..))))))))).))))))))))..... ( -33.00) >DroMoj_CAF1 58570 120 + 1 GUCGUUGUACGGCACUCCGAAAGAGGAGCCAAUGCCCAAUAUACCGAUAGUAUCGGAAAAAGUCUCGGCCAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCG (((..(((.((((....(....).(((((((..(((.......(((((...)))))..........)))(((((..((....))..)))))......))))))).))))))))))..... ( -33.13) >consensus GUCGUUGUACGGCACUCCGAAAGAGGAGCCAAUGCCCAACAUACCGAUAGUGUCGGAAAAAGUCUCGGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCG (((..(((.((((....(....).((((((.(((.........((((.....))))...........((.((((..((....))..))))))...))))))))).))))))))))..... (-28.99 = -29.27 + 0.28)



| Location | 9,806,353 – 9,806,473 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -30.35 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9806353 120 + 20766785 AUACCGAUCGUGUCGGAAAAAGUCUCUGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACGGACAACCGACUGGCCAUGAAACUGUUUGGCAGCCGAAAGGCGCUGGUC .....((((.(((((((...(((...(((.((((..((....))..))))))).((((((..((((....(((....))).)))))))))).))).)))))))(((....)))...)))) ( -35.80) >DroVir_CAF1 48060 120 + 1 AUACCGAUAGUCUCGGAAAAAGUCUCGGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCCUUGGUC ..(((((.......(((((..((....))...)))))......(((((......((((((..((((.(((.......))).))))))))))......))))).(((....))).))))). ( -36.00) >DroGri_CAF1 53103 120 + 1 AUACCAAUAGUCUCGGAAAAAGUCUCGGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGGAAGGCUUUGGUC ..(((((.......(((((..((....))...)))))......(((((......((((((..((((.(((.......))).))))))))))......)))))((((....))))))))). ( -37.40) >DroWil_CAF1 53206 120 + 1 AUACCUAUCGUGUCCGAGAAGGUCUCUGCUAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCAUUGGUC ..((((.(((....)))..))))....((((((..........(((((......((((((..((((.(((.......))).))))))))))......))))).(((....))))))))). ( -35.80) >DroMoj_CAF1 58610 120 + 1 AUACCGAUAGUAUCGGAAAAAGUCUCGGCCAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCCUUGGUC ...(((((...)))))..........((((((...........(((((......((((((..((((.(((.......))).))))))))))......))))).(((....))).)))))) ( -37.90) >DroAna_CAF1 44832 120 + 1 AUACCGAUCGUGUCGGAAAAAGUCUCUGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCAACGGACAACCGACUGGCCAUGAAACUGUUUGGCAGCCGAAAGGCACUCGUC ...(((((...)))))...........((((............(((((..(((.((((((..((((....(((....))).))))))))))...)))))))).(((....)))..)))). ( -35.20) >consensus AUACCGAUAGUGUCGGAAAAAGUCUCGGCGAAUUUCCUAAAAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCAUUGGUC ..((((........(((((..((....))...)))))......(((((......((((((..((((.(((.......))).))))))))))......))))).(((....)))..)))). (-30.35 = -31.10 + 0.75)

| Location | 9,806,393 – 9,806,513 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -31.82 |
| Energy contribution | -31.77 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9806393 120 + 20766785 AAAGUCAAUUGCAAUCAUGGUUCCAGCCGACGGACAACCGACUGGCCAUGAAACUGUUUGGCAGCCGAAAGGCGCUGGUCAAGGAGCGCAUACGUCAGAAAACUUCCGGGCACUGGGUCA ...(((...(((..((((((..((((....(((....))).))))))))))..((.((((((.(((....)))....)))))).)).)))..((..((....))..)))))......... ( -37.50) >DroVir_CAF1 48100 120 + 1 AAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCCUUGGUCAAAGAACGCAUACGUCAGAAAACUUCCGGGCAUUGGGUAA ...(((...(((..((((((..((((.(((.......))).)))))))))).....((((((.(((....)))....))))))....)))..((..((....))..)))))......... ( -34.60) >DroGri_CAF1 53143 120 + 1 AAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGGAAGGCUUUGGUCAAAGAACGCAUACGUCAGAAAACUUCCGGGCAUUGGGUUA .((.(((((.((....(((((((..((((((((((....)))))((((..........))))((((....)))))))))....)))).))).((..((....))..)).))))))).)). ( -35.70) >DroMoj_CAF1 58650 120 + 1 AAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCCUUGGUCAAAGAACGCAUACGUCAGAAAACUUCCGGGCAUUGGGUAA ...(((...(((..((((((..((((.(((.......))).)))))))))).....((((((.(((....)))....))))))....)))..((..((....))..)))))......... ( -34.60) >DroAna_CAF1 44872 120 + 1 AAAGUCAAUUGCAAUCAUGGUUCCAGCCAACGGACAACCGACUGGCCAUGAAACUGUUUGGCAGCCGAAAGGCACUCGUCAAGGAGCGAAUACGUCAGAAAACUUCCGGGCAUUGGGUCA ....(((((.((..((((((..((((....(((....))).)))))))))).....((((((.(((....)))..((((......))))....))))))..........))))))).... ( -35.70) >DroPer_CAF1 42943 120 + 1 AAAGUCAAUUGCAAUCAUGGUUCCAGCCGACGGACAAUCGUUUGGCCAUGAAACUAUUUGGCAGCCGCAAGGCAUUGGUCAAGGAGCGCAUACGUCAGAAAACUUCCGGUCAUUGGGUCA ....(((((.((....((((((((.((((((((((....)))))((((..........)))).(((....))).)))))...))))).))).((..((....))..)))).))))).... ( -33.70) >consensus AAAGUCAAUUGCAAUCAUGGUUCCAGCCGACAGACAAUCGUCUGGCCAUGAAACUAUUUGGCAGCCGCAAGGCAUUGGUCAAAGAACGCAUACGUCAGAAAACUUCCGGGCAUUGGGUCA ...(((...(((..((((((..((((.(((.......))).))))))))))..((.((((((.(((....)))....)))))).)).)))..((..((....))..)))))......... (-31.82 = -31.77 + -0.06)

| Location | 9,806,433 – 9,806,544 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -33.21 |
| Energy contribution | -32.90 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9806433 111 + 20766785 ACUGGCCAUGAAACUGUUUGGCAGCCGAAAGGCGCUGGUCAAGGAGCGCAUACGUCAGAAAACUUCCGGGCACUGGGUCAUACACCCGUGCAGUUCAUUCAGGUGAG------UUCC--- (((.(((.((((.((..(..((.(((....)))))..)...))((((.....((..((....))..)).((((.((((.....)))))))).)))).))))))).))------)...--- ( -40.30) >DroVir_CAF1 48140 111 + 1 UCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCCUUGGUCAAAGAACGCAUACGUCAGAAAACUUCCGGGCAUUGGGUAAUACACCCGUGCAGUUCAUUCAGGUAUG------CCAC--- ..((((..((((....((((((.(((....)))....))))))((((((((..(((.(........).)))...((((.....)))))))).)))).)))).....)------))).--- ( -35.10) >DroPse_CAF1 43018 114 + 1 UUUGGCCAUGAAACUAUUUGGCAGCCGCAAGGCAUUGGUCAAGGAGCGCAUACGUCAGAAAACUUCCGGUCAUUGGGUCAUACACCCGUGCAGUUCAUUCAGGUGAG------UUGUGCU ..(((((..........(((((.(((....)))....)))))((((((....))).........))))))))..((((.....))))(..((..((((....)))).------.))..). ( -35.40) >DroGri_CAF1 53183 117 + 1 UCUGGCCAUGAAAUUAUUUGGCAGCCGGAAGGCUUUGGUCAAAGAACGCAUACGUCAGAAAACUUCCGGGCAUUGGGUUAUACACCCGUGCAGUUCAUUCAGGUAAUUGAAACCCAC--- ((((((.(((......((((((((((....))))...)))))).....)))..))))))........((((((.((((.....))))))))......(((((....)))))..))..--- ( -35.80) >DroWil_CAF1 53286 108 + 1 UCUGGCCAUGAAAUUAUUUGGCAGCCGCAAGGCAUUGGUCAAAGAGCGCAUACGUCAGAAAACUUCCGGGCAUUGGGUAAUACACCCUUGCAGUUCAUUCAGGUAUU------U------ ....(((.((((....((((((.(((....)))....))))))((((.....((..((....))..)).(((..((((.....)))).))).)))).)))))))...------.------ ( -33.00) >DroPer_CAF1 42983 114 + 1 UUUGGCCAUGAAACUAUUUGGCAGCCGCAAGGCAUUGGUCAAGGAGCGCAUACGUCAGAAAACUUCCGGUCAUUGGGUCAUACACCCGUGCAGUUCAUUCAGGUGAG------UUGUGCU ..(((((..........(((((.(((....)))....)))))((((((....))).........))))))))..((((.....))))(..((..((((....)))).------.))..). ( -35.40) >consensus UCUGGCCAUGAAACUAUUUGGCAGCCGCAAGGCAUUGGUCAAAGAGCGCAUACGUCAGAAAACUUCCGGGCAUUGGGUCAUACACCCGUGCAGUUCAUUCAGGUAAG______UUAC___ ....(((.((((....((((((.(((....)))....))))))((((((((.((..((....))..))......((((.....)))))))).)))).)))))))................ (-33.21 = -32.90 + -0.31)

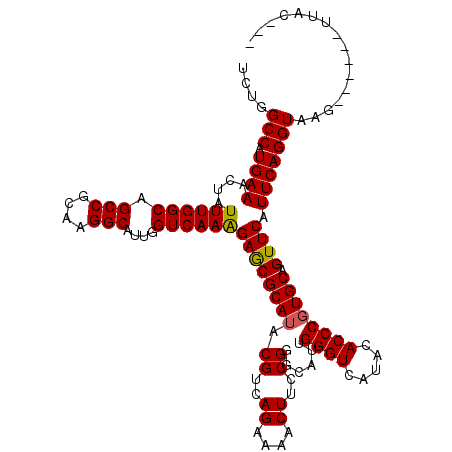
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:07 2006