| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,794,569 – 9,794,682 |
| Length | 113 |
| Max. P | 0.999788 |

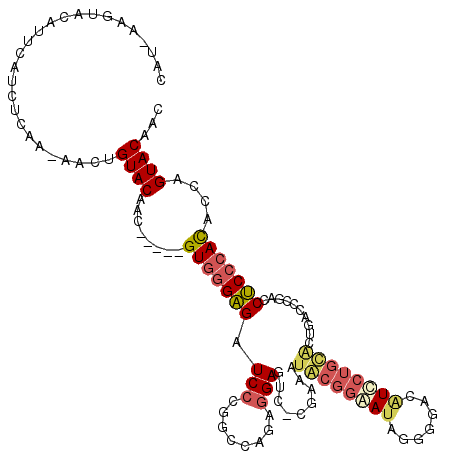
| Location | 9,794,569 – 9,794,682 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.63 |
| Mean single sequence MFE | -42.96 |
| Consensus MFE | -25.19 |
| Energy contribution | -26.83 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9794569 113 + 20766785 GUUGUACUGGUGUGGGAGGUCGGUUCAGUGCAGGACGUCCGUUAUCCCGCAUUUCG-GACUCCUCUGGCCGGGAUCUCCCAC-----GUUGUACAGUU-UUGAGAUGAAUGUACUUAAUG .((((((...((((((((((((((.(((...((((.(((((..((.....))..))-)))))))))))))..))))))))))-----)..))))))..-(((((((....)).))))).. ( -42.10) >DroSec_CAF1 32766 113 + 1 GUUGUACUGGUGUGGGAGAUCGGGUCAGUGCAGGAUGUCCCCUAUUCCGCAUAUCG-GACUCCUCUGGCCGGGAUCUCCCAC-----GUUGUACAGUU-UUGAGAUGAAUGUACCUAAUG .((((((...(((((((((((.((((((...((((.((((..(((.....)))..)-)))))))))))))..))))))))))-----)..))))))..-..................... ( -43.70) >DroEre_CAF1 35715 112 + 1 GUUGUACUGGCGUGGGAGGUCAGGUCCGUACAGGAUGUCCCUUAUUCCGUAUUUCG-GACUCCUCUGGCCGGGAUCUCCCAC-----UCAGUACAGUU-UUGAGAUGAAUGUACUU-AUG .(((((((((.((((((((((.((.(((...((((.((((...((.....))...)-))))))).)))))..))))))))))-----)))))))))..-.................-... ( -44.80) >DroYak_CAF1 34192 113 + 1 GUUGUACUGGCGUGGGAGGUCGGGUCAGCACUGGAUGUCCCCUAUCCCGUAUUUCGGGACUCCUCUGACCAGGAUCUCCCAC-----GUUGUACAGUU-UUGAGAUGAAUGUACUU-UUG .((((((.(((((((((((((.((((((....(((.(((((..(........)..)))))))).))))))..))))))))))-----)))))))))..-.................-... ( -50.40) >DroAna_CAF1 33113 107 + 1 CCUGUACUGUUAUCGGGGGUGCG--------AGAAAAAUCCCUAUUUCUUGUUUAU-GUGUC--AUGAUGGGGAUCCCUCACACCCCUUUGUACGGUUUUUGAGAUGAAUGUU--UUAUG .((((((.......(((((((.(--------((....(((((((((...((.....-....)--).)))))))))..))).)))))))..))))))....((((((....)))--))).. ( -33.80) >consensus GUUGUACUGGUGUGGGAGGUCGGGUCAGUACAGGAUGUCCCCUAUUCCGUAUUUCG_GACUCCUCUGGCCGGGAUCUCCCAC_____GUUGUACAGUU_UUGAGAUGAAUGUACUU_AUG .((((((....((((((((((.((((((...((((.(((..................)))))))))))))..))))))))))........))))))........................ (-25.19 = -26.83 + 1.64)

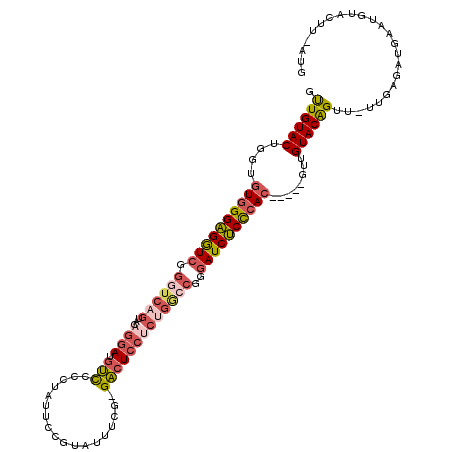

| Location | 9,794,569 – 9,794,682 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.63 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.74 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.45 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9794569 113 - 20766785 CAUUAAGUACAUUCAUCUCAA-AACUGUACAAC-----GUGGGAGAUCCCGGCCAGAGGAGUC-CGAAAUGCGGGAUAACGGACGUCCUGCACUGAACCGACCUCCCACACCAGUACAAC .....................-...(((((...-----(((((((....(((.(((.((...)-)....((((((((.......)))))))))))..)))..)))))))....))))).. ( -32.80) >DroSec_CAF1 32766 113 - 1 CAUUAGGUACAUUCAUCUCAA-AACUGUACAAC-----GUGGGAGAUCCCGGCCAGAGGAGUC-CGAUAUGCGGAAUAGGGGACAUCCUGCACUGACCCGAUCUCCCACACCAGUACAAC .....................-...(((((...-----((((((((((..((.((((((((((-(..(((.....)))..)))).))))...))).)).))))))))))....))))).. ( -39.40) >DroEre_CAF1 35715 112 - 1 CAU-AAGUACAUUCAUCUCAA-AACUGUACUGA-----GUGGGAGAUCCCGGCCAGAGGAGUC-CGAAAUACGGAAUAAGGGACAUCCUGUACGGACCUGACCUCCCACGCCAGUACAAC ...-.................-...(((((((.-----(((((((.((..(((((.(((((((-(...............)))).)))).)..)).)).)).)))))))..))))))).. ( -38.26) >DroYak_CAF1 34192 113 - 1 CAA-AAGUACAUUCAUCUCAA-AACUGUACAAC-----GUGGGAGAUCCUGGUCAGAGGAGUCCCGAAAUACGGGAUAGGGGACAUCCAGUGCUGACCCGACCUCCCACGCCAGUACAAC ...-.................-...(((((..(-----(((((((.((..((((((.(((((((((.....)))))).(....).)))....)))))).)).))))))))...))))).. ( -44.90) >DroAna_CAF1 33113 107 - 1 CAUAA--AACAUUCAUCUCAAAAACCGUACAAAGGGGUGUGAGGGAUCCCCAUCAU--GACAC-AUAAACAAGAAAUAGGGAUUUUUCU--------CGCACCCCCGAUAACAGUACAGG .....--.................((((((...(((((((((((((((((..((.(--(....-.....)).))....))))))..)))--------)))))))).(....).)))).)) ( -30.00) >consensus CAU_AAGUACAUUCAUCUCAA_AACUGUACAAC_____GUGGGAGAUCCCGGCCAGAGGAGUC_CGAAAUACGGAAUAGGGGACAUCCUGCACUGACCCGACCUCCCACACCAGUACAAC ..........................((((........(((((((.(((........))).........((((((((.......))))))))..........)))))))....))))... (-16.74 = -17.74 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:58 2006