| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,751,537 – 9,751,643 |
| Length | 106 |
| Max. P | 0.825947 |

| Location | 9,751,537 – 9,751,643 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -20.31 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
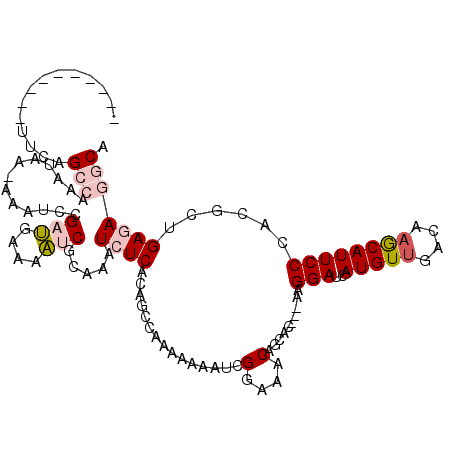
>2R_DroMel_CAF1 9751537 106 - 20766785 ----------UACAGCCAAAUAACAAAUCCGAUGAAAAUCGCAAAUCUCACAGCCAAAAAAAUCGGAAACAGCACAACAAGGAUCAUGUUGAUAAGCAUUCCCACGCUGAGAGGCA ----------....(((............((((....))))......((.((((..........(....)..........(((..(((((....))))))))...)))).))))). ( -23.10) >DroVir_CAF1 140 110 - 1 UUAUUACUUUUCAAGCCAAAU---AAGUCGGAGGAAACACGCAAAUCUCACACACAAAAGAAUAGGAAACAGCAU---AAGGAACAUGUUGAUAAACAUUCCCAAGCGGAAAAACA .......(((((.........---........(....).(((......................(....).....---..(((..(((((....))))))))...))))))))... ( -15.90) >DroGri_CAF1 569 103 - 1 CA----------UAGCCAAAU---AAAUCGGAGGAAACACGCAAAUCUCACACUCAAAAGAAUAGGAAGCAUCAUAGUAAGGACCAUGUUGAUAAACAUUCCCAAGCGGAAAAACA ..----------.........---...((...(....).(((.....((....((....))....)).............(((..(((((....))))))))...)))))...... ( -10.90) >DroSim_CAF1 608 106 - 1 ----------UUCAGCCAAAUAACAAAUCCGAUGAAAAUCGCAAAUCUCACAGCCAAAAAAAUCGGAAACAGCACAACAAGGAUCAUGUUGACAAGCAUUCCCACGCUGAGAGGCA ----------....(((............((((....))))......((.((((..........(....)..........(((..(((((....))))))))...)))).))))). ( -23.10) >DroEre_CAF1 611 103 - 1 ----------UUUAGCCAAAUAAUAAAUCCGAUGAAAAUCGCAAAUCUCACAGCCAAAAAAGUCGGAAACAGCAC---AAGGAUCAUGUUGACAAGCAUUCCCACGCUGAGAGGCA ----------....(((..........((((((.......((..........)).......))))))..((((..---..(((..(((((....))))))))...))))...))). ( -24.84) >DroYak_CAF1 611 103 - 1 ----------UUCAGCCAAAUAAUAAAUCCGAUGAAAAUCGCAAAUCUCACAGCCAAAAAAGUCGGAAACAGCAC---AAGGAUCAUGUGGACAAGCAUUCCCACGCUGAGAGGCA ----------....(((............((((....))))....((((............((((....)..(((---(.......))))))).(((........)))))))))). ( -24.00) >consensus __________UUCAGCCAAAUAA_AAAUCCGAUGAAAAUCGCAAAUCUCACAGCCAAAAAAAUCGGAAACAGCAC___AAGGAUCAUGUUGACAAGCAUUCCCACGCUGAGAGGCA ..............(((.............(((....))).....((((...............(....)..........(((..(((((....))))))))......))))))). (-13.52 = -14.35 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:48 2006