| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
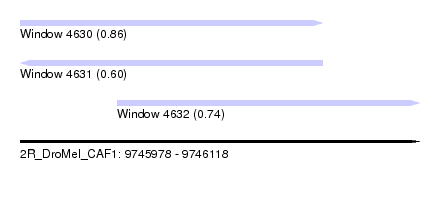
| Location | 9,745,978 – 9,746,118 |
| Length | 140 |
| Max. P | 0.861207 |

| Location | 9,745,978 – 9,746,084 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.34 |
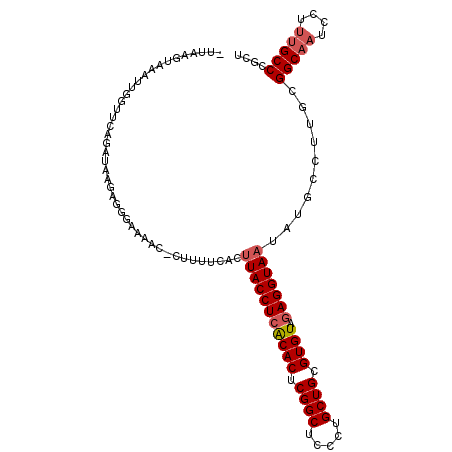
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
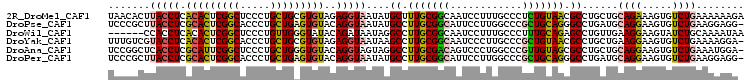
>2R_DroMel_CAF1 9745978 106 + 20766785 -UUAAGUAAAUUGGUUCAGAUAAGAGGGAAAAC-CUUAACACUUACCUCACACUCGGCUCCCUGCUGCGUGUAGAGGUAAUAUGCUUUGCGGCAAUCCUUUGCCCUCU -....(((((.............((((.....)-))).....(((((((((((.((((.....)))).)))).))))))).....)))))(((((....))))).... ( -33.60) >DroPse_CAF1 14650 107 + 1 -UUAUAUAUAUAUAAUGAGAUUGAAUGUGAUAUGUUUCCCGCUUACCUCGCACUCGGCACCCUGCUGAGUGUACAGGUAAUAUGCCUUGCGGCAUUCCUUGGCCCGCU -....((((((...((........))...)))))).....((((((((.(((((((((.....)))))))))..)))))).(((((....)))))..........)). ( -29.40) >DroSec_CAF1 3723 106 + 1 -UUAGCUAAAUUGGUUCAGGUUAGAGGGAAAAC-CUUUUCCCUUACCUCACACUCGGCUCCCUGCUGCGUGUAGAGGUAAUAUGCCUUUCGGCAAUCCUUUGCCCGCU -..(((......(((...((..(((((.....)-))))..))(((((((((((.((((.....)))).)))).)))))))...)))....(((((....))))).))) ( -39.90) >DroSim_CAF1 3716 106 + 1 -UUAGCUAAAUUGGUUCCGAUAAGAGGGAAAAC-CUUUUCACUUACCUCACACUCGGCUCCCUGCUGCGUGUAGAGGUAAUAUGCCUUUCGGCAAUCCUUUGCCCGCU -..(((......(((......((((((.....)-)))))...(((((((((((.((((.....)))).)))).)))))))...)))....(((((....))))).))) ( -35.90) >DroEre_CAF1 3619 106 + 1 AUUAAGUAAGUUGGUUCAGCUAAGCG-UAAAAC-CUUUUGCCUUACCUCACACUCGGCUCCCUGCUGCGUGUAGAGGUAAUAGGCCUUGCGGCAAUCCUUUGCCCGCU .....(((((..((((..((.....)-)..)))-)....((((((((((((((.((((.....)))).)))).))))))..)))))))))(((((....))))).... ( -39.80) >DroYak_CAF1 3740 106 + 1 UUCAAGUAAAUUGGUUCAGAUAAGCG-UAAAAC-CUUUUGUCGUACCUCACACUCGGCACCCUGCUGCGUGUAGAGGUAAUAAGCCUUGCGGCAAUCCCUUGCCCGCU ............((((..((((((.(-......-).)))))).((((((((((.((((.....)))).)))).))))))...))))..(((((((....))).)))). ( -32.90) >consensus _UUAAGUAAAUUGGUUCAGAUAAGAGGGAAAAC_CUUUUCACUUACCUCACACUCGGCUCCCUGCUGCGUGUAGAGGUAAUAUGCCUUGCGGCAAUCCUUUGCCCGCU ..........................................(((((((((((.((((.....)))).)))).)))))))..........(((((....))))).... (-24.35 = -24.88 + 0.53)

| Location | 9,745,978 – 9,746,084 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.34 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -22.59 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9745978 106 - 20766785 AGAGGGCAAAGGAUUGCCGCAAAGCAUAUUACCUCUACACGCAGCAGGGAGCCGAGUGUGAGGUAAGUGUUAAG-GUUUUCCCUCUUAUCUGAACCAAUUUACUUAA- .(((((....(((..(((....(((((.(((((((.((((.(.((.....)).).))))))))))))))))..)-))..))))))))....................- ( -31.40) >DroPse_CAF1 14650 107 - 1 AGCGGGCCAAGGAAUGCCGCAAGGCAUAUUACCUGUACACUCAGCAGGGUGCCGAGUGCGAGGUAAGCGGGAAACAUAUCACAUUCAAUCUCAUUAUAUAUAUAUAA- .((..(((.(((.(((((....)))))....)))...(((((.((.....)).)))))...)))..)).(....)................................- ( -30.20) >DroSec_CAF1 3723 106 - 1 AGCGGGCAAAGGAUUGCCGAAAGGCAUAUUACCUCUACACGCAGCAGGGAGCCGAGUGUGAGGUAAGGGAAAAG-GUUUUCCCUCUAACCUGAACCAAUUUAGCUAA- (((.((...(((..((((....))))..(((((((.((((.(.((.....)).).)))))))))))((((((..-..)))))).....)))...))......)))..- ( -39.10) >DroSim_CAF1 3716 106 - 1 AGCGGGCAAAGGAUUGCCGAAAGGCAUAUUACCUCUACACGCAGCAGGGAGCCGAGUGUGAGGUAAGUGAAAAG-GUUUUCCCUCUUAUCGGAACCAAUUUAGCUAA- (((..((.......((((....))))..(((((((.((((.(.((.....)).).))))))))))))).....(-((..(((........))))))......)))..- ( -33.30) >DroEre_CAF1 3619 106 - 1 AGCGGGCAAAGGAUUGCCGCAAGGCCUAUUACCUCUACACGCAGCAGGGAGCCGAGUGUGAGGUAAGGCAAAAG-GUUUUA-CGCUUAGCUGAACCAACUUACUUAAU .((((.(((....)))))))(((((((..((((((.((((.(.((.....)).).))))))))))))))....(-((((..-(.....)..)))))..)))....... ( -36.00) >DroYak_CAF1 3740 106 - 1 AGCGGGCAAGGGAUUGCCGCAAGGCUUAUUACCUCUACACGCAGCAGGGUGCCGAGUGUGAGGUACGACAAAAG-GUUUUA-CGCUUAUCUGAACCAAUUUACUUGAA ......((((((((((((....)))....((((((.((((.(.((.....)).).))))))))))........(-((((..-.........)))))))))).)))).. ( -30.30) >consensus AGCGGGCAAAGGAUUGCCGCAAGGCAUAUUACCUCUACACGCAGCAGGGAGCCGAGUGUGAGGUAAGGGAAAAG_GUUUUACCUCUUAUCUGAACCAAUUUACUUAA_ ...((....(((...(((....)))...(((((((.((((.(.((.....)).).)))))))))))......................)))...))............ (-22.59 = -23.40 + 0.81)



| Location | 9,746,012 – 9,746,118 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9746012 106 + 20766785 UAACACUUACCUCACACUCGGCUCCCUGCUGCGUGUAGAGGUAAUAUGCUUUGCGGCAAUCCUUUGCCCUCUGUAACGCCUGCUGCAGAAAGUGUCUGAAAAAAGA ..(((((((((((((((.((((.....)))).)))).))))).......(((((((((...(.((((.....)))).)..)))))))))))))))........... ( -36.10) >DroPse_CAF1 14685 105 + 1 UCCCGCUUACCUCGCACUCGGCACCCUGCUGAGUGUACAGGUAAUAUGCCUUGCGGCAUUCCUUGGCCCGCUGCAGGGCCUGAUGCAGGAAGUGUCUGAAGGAGG- ..((..((((((.(((((((((.....)))))))))..)))))).(((((....)))))((((((((((......))))).(((((.....)))))..)))))))- ( -46.30) >DroWil_CAF1 3790 100 + 1 ------CCACCUCACACUCGGCUCCCUGUUGGGUAUACAGAUAAUAGGCCUUGCGGCAAUCCUUUGCCCUUUGCAGAGCCUGUUGAAGGAAGUAUCUGCAAAAUAA ------...(((...(((((((.....))))))).......((((((((.(((((((((....)))))....)))).)))))))).)))................. ( -29.70) >DroYak_CAF1 3774 105 + 1 UUUGUCGUACCUCACACUCGGCACCCUGCUGCGUGUAGAGGUAAUAAGCCUUGCGGCAAUCCCUUGCCCGCUGUAACGCCUGCUGAAGGAAGUGUCUGAAAAGGA- .......((((((((((.((((.....)))).)))).)))))).....((((..(((((....)))))((((......(((.....))).))))......)))).- ( -33.60) >DroAna_CAF1 3946 105 + 1 UCCGGCUCACCUCGCAUUCGGCUCCCUGCUGGGUGUACAGGUAGUAGGCCUUGCGACAGUCCCUGGCCCGUUGUAGCGCCUGCUGCAGGAAGUGUCUGAAAUGGA- ((((.(.(((.(((((((((((.....)))))))))....(((((((((.(((((((.((.....))..))))))).)))))))))..)).)))...)...))))- ( -43.00) >DroPer_CAF1 13689 105 + 1 UCCCGCUUACCUCGCACUCGGCACCCUGCUGAGUGUACAGGUAAUAUGCCUUGCGGCAUUCCUUGGCCCGCUGCAGGGCCUGAUGCAGGAAGUGUCUGAAGGAGG- ..((..((((((.(((((((((.....)))))))))..)))))).(((((....)))))((((((((((......))))).(((((.....)))))..)))))))- ( -46.30) >consensus UCCCGCUUACCUCACACUCGGCACCCUGCUGAGUGUACAGGUAAUAUGCCUUGCGGCAAUCCUUGGCCCGCUGCAGCGCCUGCUGCAGGAAGUGUCUGAAAAAGA_ .......(((((.(((((((((.....)))))))))..)))))....((.(((((((............))))))).))......((((.....))))........ (-22.64 = -22.70 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:46 2006