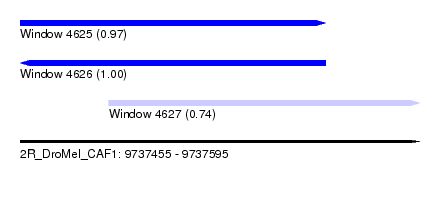
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,737,455 – 9,737,595 |
| Length | 140 |
| Max. P | 0.995677 |

| Location | 9,737,455 – 9,737,562 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -24.96 |
| Energy contribution | -26.27 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9737455 107 + 20766785 UUUGCUUAGCAGCUCGUUUCGGGGUCGA-------AUCGAAAUCGGAAUCAGAAUCGUAUUGAGAUUUUUAUUCGGAUUGGGAUUGGAAACAGUGCCCGAAACGGAGAGAUGGG ....((((....(((((((((((....(-------(((..((((.((((.((((((.......)))))).)))).))))..))))(....)....)))))))).)))...)))) ( -34.90) >DroSec_CAF1 6428 113 + 1 UUUGCUUAGCAGCUCGUUUCGGGGUCGAGUU-GGAAUCGGAAUCGAAAUCGGAAUCGUAUUGAGAUUUUUAUUCGGAUUGGGUUUGGAAACAGUGCCCGAAUCGGAGAGAUGGG .((((...))))((((((((....((((.((-.((((..(((((..(((((....)).)))..)))))..)))).))(((((((((....))).)))))).)))).)))))))) ( -34.20) >DroSim_CAF1 6491 113 + 1 UUUGCUUAGCAGCUCGUUUCGGGGUCGAAUC-GGAAUCGAAAUCGGAAUCGGAAUCGUAUUGAGAUUUGUAUUCGGAUUGGGUUUGGAAACAGUGCCCGAAACGGAGAGAUGGG .((((...))))((((((((.....((((((-..((((((..(((....)))..))).)))..))))))..((((..(((((((((....))).))))))..)))))))))))) ( -35.00) >DroYak_CAF1 6356 104 + 1 ----------AGCUCGCUUCGGGGUUGAAUCCGGAAUCUGAAUCUGAAUCGGAAUCUGAAUCAGAAUCAGAAUCGUAUUCGGAUUGGAAACAGAGCCCGAAACGGAGAGAUGGG ----------..((((.((((((.((.((((((((.(((((.(((((.((((...)))).))))).)))))......))))))))(....).)).)))))).).)))....... ( -37.50) >consensus UUUGCUUAGCAGCUCGUUUCGGGGUCGAAUC_GGAAUCGAAAUCGGAAUCGGAAUCGUAUUGAGAUUUUUAUUCGGAUUGGGAUUGGAAACAGUGCCCGAAACGGAGAGAUGGG ............(((((((((((.((((........))))((((.((((..(((((.......)))))..)))).))))....(((....)))..)))))))).)))....... (-24.96 = -26.27 + 1.31)

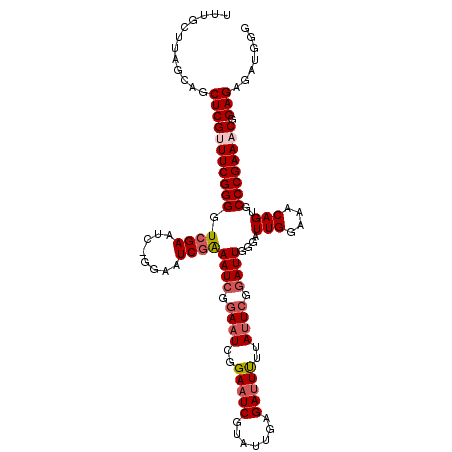
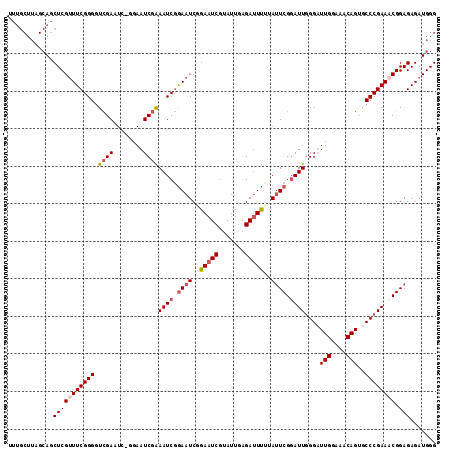
| Location | 9,737,455 – 9,737,562 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.95 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9737455 107 - 20766785 CCCAUCUCUCCGUUUCGGGCACUGUUUCCAAUCCCAAUCCGAAUAAAAAUCUCAAUACGAUUCUGAUUCCGAUUUCGAU-------UCGACCCCGAAACGAGCUGCUAAGCAAA ..........(((((((((....(((...((((..((((.((((...((((.......))))...)))).))))..)))-------).)))))))))))).(((....)))... ( -26.40) >DroSec_CAF1 6428 113 - 1 CCCAUCUCUCCGAUUCGGGCACUGUUUCCAAACCCAAUCCGAAUAAAAAUCUCAAUACGAUUCCGAUUUCGAUUCCGAUUCC-AACUCGACCCCGAAACGAGCUGCUAAGCAAA ..........((.((((((....(((....)))..((((.((((..(((((.............)))))..)))).))))..-........)))))).)).(((....)))... ( -19.82) >DroSim_CAF1 6491 113 - 1 CCCAUCUCUCCGUUUCGGGCACUGUUUCCAAACCCAAUCCGAAUACAAAUCUCAAUACGAUUCCGAUUCCGAUUUCGAUUCC-GAUUCGACCCCGAAACGAGCUGCUAAGCAAA ..........(((((((((................((((.((((...((((.......))))...)))).))))((((....-...)))).))))))))).(((....)))... ( -27.00) >DroYak_CAF1 6356 104 - 1 CCCAUCUCUCCGUUUCGGGCUCUGUUUCCAAUCCGAAUACGAUUCUGAUUCUGAUUCAGAUUCCGAUUCAGAUUCAGAUUCCGGAUUCAACCCCGAAGCGAGCU---------- ..........(((((((((....(((...((((((.....((.(((((.(((((.((.......)).))))).))))).)))))))).))))))))))))....---------- ( -35.50) >consensus CCCAUCUCUCCGUUUCGGGCACUGUUUCCAAACCCAAUCCGAAUAAAAAUCUCAAUACGAUUCCGAUUCCGAUUCCGAUUCC_GAUUCGACCCCGAAACGAGCUGCUAAGCAAA ..........(((((((((................((((.((((...((((.......))))...)))).)))).(((........)))..))))))))).(((....)))... (-17.26 = -18.95 + 1.69)

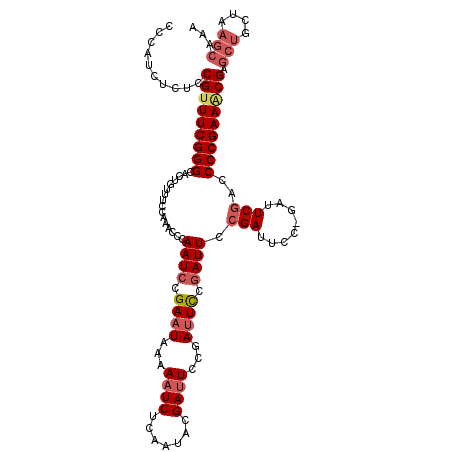
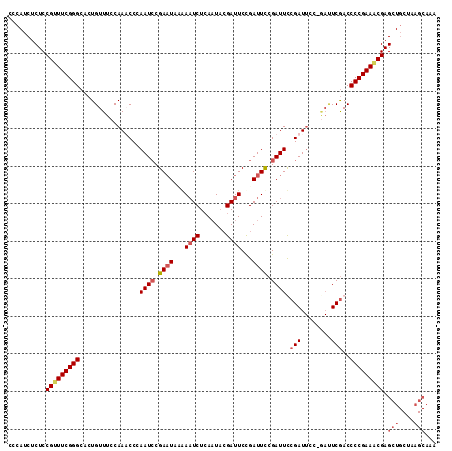
| Location | 9,737,486 – 9,737,595 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -18.69 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
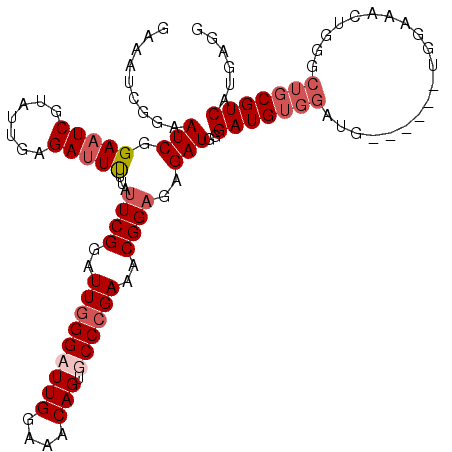
>2R_DroMel_CAF1 9737486 109 + 20766785 GAAAUCGGAAUCAGAAUCGUAUUGAGAUUUUUAUUCGGAUUGGGAUUGGAAACAGUGCCCGAAACGGAGAGAUGGGAUGUGGAUG------UGGAAACUGGGCUGCGUCAUGAGG ..((((.((((.((((((.......)))))).)))).))))(((((((....)))).)))...............((((..(...------.(....)....)..))))...... ( -33.00) >DroSec_CAF1 6465 103 + 1 GGAAUCGAAAUCGGAAUCGUAUUGAGAUUUUUAUUCGGAUUGGGUUUGGAAACAGUGCCCGAAUCGGAGAGAUGGGAUG------------UGGAAACUGGGCUGCGUCAUGAGG ((((((..(((((....)).)))..))))))..(((((.(((((((((....))).)))))).))))).......((((------------..(........)..))))...... ( -28.00) >DroSim_CAF1 6528 109 + 1 GAAAUCGGAAUCGGAAUCGUAUUGAGAUUUGUAUUCGGAUUGGGUUUGGAAACAGUGCCCGAAACGGAGAGAUGGGAUGUGGCUG------UGGAAACUGGGCUGCGUCAUGAGG ....(((...((.(((((.......)))))...((((..(((((((((....))).))))))..))))))..)))((((..(((.------.(....)..)))..))))...... ( -36.10) >DroYak_CAF1 6384 115 + 1 UGAAUCUGAAUCGGAAUCUGAAUCAGAAUCAGAAUCGUAUUCGGAUUGGAAACAGAGCCCGAAACGGAGAGAUGGGAUGUGGAUGUUGAUGUGAAAAGUGGGCUGCGUCAUGAGG (((.(((((.((((...)))).))))).)))...((((.(((((.(((....)))...)))))))))........(((((....(((.((.......)).))).)))))...... ( -28.30) >consensus GAAAUCGGAAUCGGAAUCGUAUUGAGAUUUUUAUUCGGAUUGGGAUUGGAAACAGUGCCCGAAACGGAGAGAUGGGAUGUGGAUG______UGGAAACUGGGCUGCGUCAUGAGG .........(((.(((((.......)))))...((((..(((((((((....))).))))))..))))..)))..(((((((....................)))))))...... (-18.69 = -20.50 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:41 2006