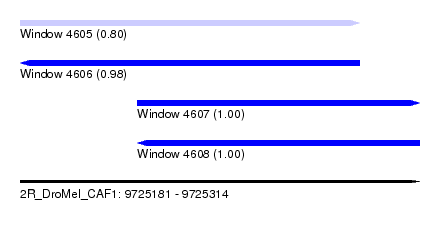
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,725,181 – 9,725,314 |
| Length | 133 |
| Max. P | 0.999735 |

| Location | 9,725,181 – 9,725,294 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
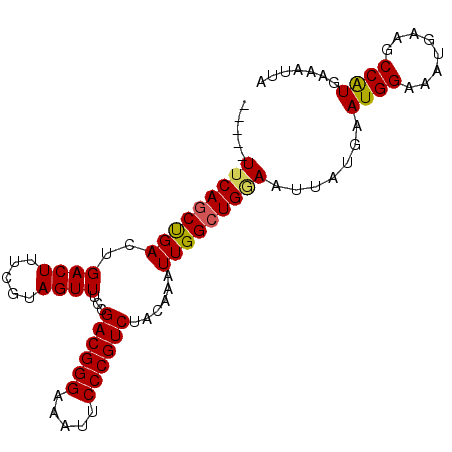
>2R_DroMel_CAF1 9725181 113 + 20766785 GUGUGCGGACUUAUCAGAGC-UUUUCAUCUGUACUUAAUUUAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA .((.(((((.((((((((..-......))))................((((........))))..)))).))).)))).((((((((((((.....))))))(....))))))) ( -26.80) >DroSec_CAF1 43312 105 + 1 --------ACUUAUCAGAGC-UUUUCAUCUGUACUUAGUUUAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA --------(((...((((..-......)))).....))).........(((((....................))))).((((((((((((.....))))))(....))))))) ( -23.75) >DroSim_CAF1 34585 105 + 1 --------ACUUAUCAGAGC-UUUUCAUCUGUACUUAAUUUAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA --------......((((..-......)))).................(((((....................))))).((((((((((((.....))))))(....))))))) ( -23.15) >DroEre_CAF1 36065 111 + 1 G--UGCGGACUUAUCAGAGC-UUUUCAUCUGUACUUAAUUUAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUUCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA (--(((((((((....))).-......)))))))..............(((((....................))))).((((((((((((.....))))))(....))))))) ( -26.36) >DroYak_CAF1 42229 111 + 1 G--UGCGGACUUAUCAGAGC-UUUUCAUCUGUACUUAAUUUAAUUUCACGGCUUCAUUUCCAUUCAUAAUUUCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA (--(((((((((....))).-......)))))))...............((((....................))))..((((((((((((.....))))))(....))))))) ( -25.66) >DroAna_CAF1 40586 112 + 1 G--UUCGGAUUUAUCAAAGCGUUUUCAUCUGUACUUAAUUUAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA .--((((((....)).....((((((......................(((((....................))))).......((((((.....))))))))))))..)))) ( -23.45) >consensus G__UGCGGACUUAUCAGAGC_UUUUCAUCUGUACUUAAUUUAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAA ..............((((.........)))).................(((((....................))))).((((((((((((.....))))))(....))))))) (-20.73 = -21.07 + 0.33)
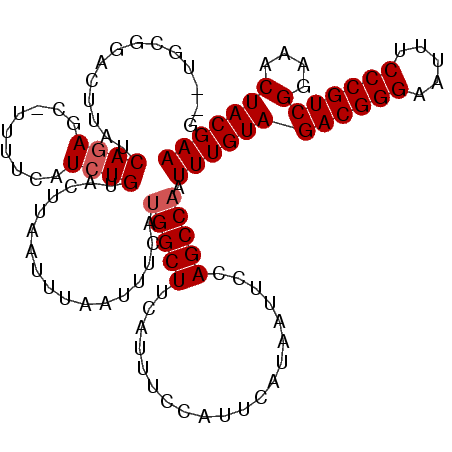
| Location | 9,725,181 – 9,725,294 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.27 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9725181 113 - 20766785 UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCUGAUAAGUCCGCACAC ....((((((..((((((.....))))))...))))))((.(((.((((..((((........))))................((((......-..)))))))).))))).... ( -25.00) >DroSec_CAF1 43312 105 - 1 UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAACUAAGUACAGAUGAAAA-GCUCUGAUAAGU-------- ....(((((((.((((((.....)))))).......(((((...(((.........)))..))))).))))))).........((((......-..))))......-------- ( -24.50) >DroSim_CAF1 34585 105 - 1 UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCUGAUAAGU-------- ....(((((((.((((((.....)))))).......(((((...(((.........)))..))))).))))))).........((((......-..))))......-------- ( -24.50) >DroEre_CAF1 36065 111 - 1 UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCUGAUAAGUCCGCA--C (((((((((((.((((((.....))))))..(.....)...))))))))))).(((.(((....)))................((((......-..)))).....)))...--. ( -25.30) >DroYak_CAF1 42229 111 - 1 UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCGUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCUGAUAAGUCCGCA--C (((((((((((.((((((.....))))))..(.....)...))))))))))).(((.(((....)))................((((......-..)))).....)))...--. ( -24.90) >DroAna_CAF1 40586 112 - 1 UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAACGCUUUGAUAAAUCCGAA--C (((((((((((.((((((.....)))))).......(((((...(((.........)))..))))).))))))).........((((((....)).)))).......))))--. ( -23.70) >consensus UUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA_GCUCUGAUAAGUCCGCA__C .((((((((((.((((((.....))))))..(.....)...))))))))))((((........))))................((((.(......))))).............. (-23.76 = -23.27 + -0.50)

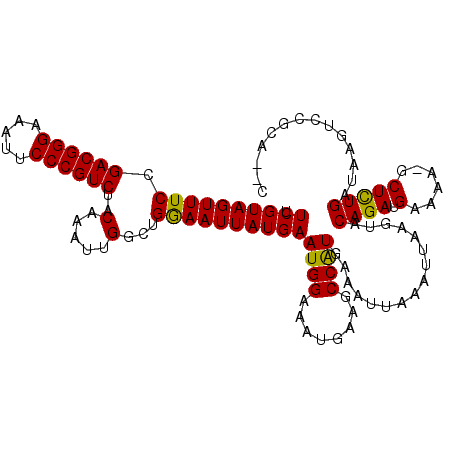
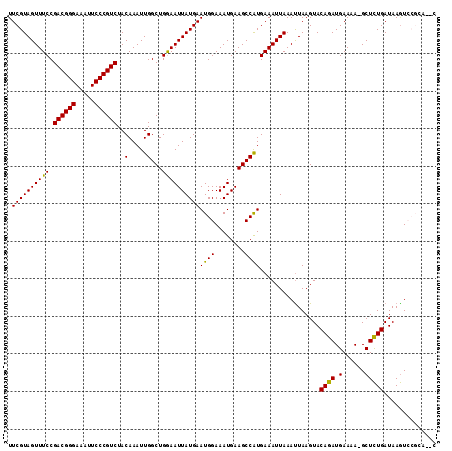
| Location | 9,725,220 – 9,725,314 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -21.65 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9725220 94 + 20766785 UAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAAAUGGC .......((((........))))...........((((.((((((((((((.....))))))(....)))))))..((((....))))..)))) ( -26.80) >DroSec_CAF1 43343 89 + 1 UAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA----- ........(((((....................))))).((((((((((((.....))))))(....)))))))..((((....)))).----- ( -24.85) >DroSim_CAF1 34616 89 + 1 UAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA----- ........(((((....................))))).((((((((((((.....))))))(....)))))))..((((....)))).----- ( -24.85) >DroEre_CAF1 36102 89 + 1 UAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUUCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA----- ........(((((....................))))).((((((((((((.....))))))(....)))))))..((((....)))).----- ( -24.85) >DroYak_CAF1 42266 89 + 1 UAAUUUCACGGCUUCAUUUCCAUUCAUAAUUUCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAUUCAGCUGAA----- ........(((((...(((((.............((......)).((((((.....)))))))))))..((....)).....)))))..----- ( -23.10) >DroAna_CAF1 40624 89 + 1 UAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCGGGUGUA----- ....................(((((....((((.((......)).((((((.....))))))))))(((((....)).))).)))))..----- ( -22.30) >consensus UAAUUUCAUGGCUUCAUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA_____ ........(((((....................))))).((((((((((((.....))))))(....)))))))..((((....))))...... (-21.65 = -22.32 + 0.67)

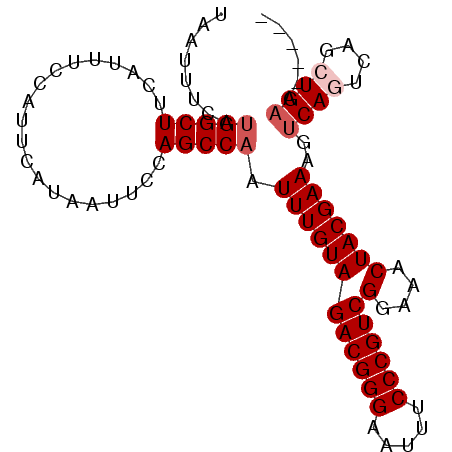
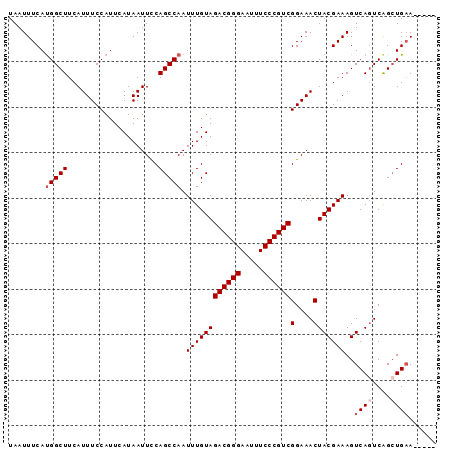
| Location | 9,725,220 – 9,725,314 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -23.79 |
| Energy contribution | -23.68 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9725220 94 - 20766785 GCCAUUUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUA ...((((((((..(..((((.....))))...((((((.....))))))......)..)))))))).....((((........))))....... ( -26.40) >DroSec_CAF1 43343 89 - 1 -----UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUA -----((((((..(..((((.....))))...((((((.....))))))......)..)))))).......((((........))))....... ( -24.80) >DroSim_CAF1 34616 89 - 1 -----UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUA -----((((((..(..((((.....))))...((((((.....))))))......)..)))))).......((((........))))....... ( -24.80) >DroEre_CAF1 36102 89 - 1 -----UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUA -----((((((..(..((((.....))))...((((((.....))))))......)..)))))).......((((........))))....... ( -25.30) >DroYak_CAF1 42266 89 - 1 -----UUCAGCUGAAUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCGUGAAAUUA -----(((((((..((..(.(((((((((((.((((((.....))))))..(.....)...))))))))))).)...))..)))..)))).... ( -25.80) >DroAna_CAF1 40624 89 - 1 -----UACACCCGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUA -----................((((((((((.((((((.....))))))..(.....)...))))))))))((((........))))....... ( -21.20) >consensus _____UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUA .....(((((((((..((((.....))))...((((((.....))))))......))))))))).......((((........))))....... (-23.79 = -23.68 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:23 2006