| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,722,313 – 9,722,454 |
| Length | 141 |
| Max. P | 0.769984 |

| Location | 9,722,313 – 9,722,415 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.42 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -25.32 |
| Energy contribution | -26.40 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
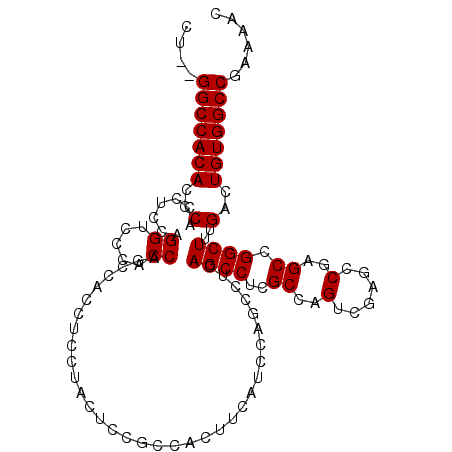
>2R_DroMel_CAF1 9722313 102 + 20766785 AGCGGUGCGGCAAGGUAAUAAAAGCCAUUUUGGUUUUCGGCCACAGUCAAAGCCGGCUCGGCUCGACUGGCGAGGCUGAAUCUGGAUAAAGUGGCGGAGUAG .((..(((.((..(((.......)))..((..(..(((((((.(((((..(((((...))))).)))))....))))))).)..))....)).)))..)).. ( -36.20) >DroSec_CAF1 40064 102 + 1 AGCGGUGCGGCAAGGUAAUAAAAGCCAUUUUGGUUUUCGGCCACAGUCAAAGCCGGCUCGGUUCGACUGGCGAGGCUGAGGCUGGAUGAAGUGGCGGAGUAG .((......))............((((((((....(((((((.(((((...(((((.(((...))))))))..))))).))))))).))))))))....... ( -39.70) >DroSim_CAF1 31340 102 + 1 AGCGGAGCGGCAAGGUAAUAAAAGCCAUUUUGGUUUUCGGCCACAGUCAAAGCCGGCUCGGCUCGACUGGCGAGGCUGAGGCUGGAUGAAGUGGCGGAGUAG .((......))............((((((((....(((((((.(((((...(((((.(((...))))))))..))))).))))))).))))))))....... ( -39.70) >DroEre_CAF1 32963 91 + 1 AGCGGUGCGGCAAGGUAAUAAAAGCCAUUUUGGUUUUCGGCCACAGUCGAAGCCAGCUC----------GCGAGGCUGGG-AAGGAUGAUGUGGCGGAGUAG .((..(((.(((.(((....((((((.....))))))..))).((..(....(((((.(----------....)))))).-..)..)).))).)))..)).. ( -29.60) >DroYak_CAF1 38870 91 + 1 AGCGGUGUGGCAAGGUAAUAAAAGCCAUUUUGGUUUUCGGCCACAGUCGAAGUCAGUUU----------GCGAGGCUGGA-AAGGAUGAGGUGGCGGAGUAG ..(((.(((((............))))).)))...(((.(((((.(((....((((((.----------....)))))).-...)))...))))).)))... ( -26.80) >consensus AGCGGUGCGGCAAGGUAAUAAAAGCCAUUUUGGUUUUCGGCCACAGUCAAAGCCGGCUCGG_UCGACUGGCGAGGCUGAG_CUGGAUGAAGUGGCGGAGUAG .((..(((.((..(((....((((((.....))))))..))).(((((...(((((..........)))))..)))))............)).)))..)).. (-25.32 = -26.40 + 1.08)


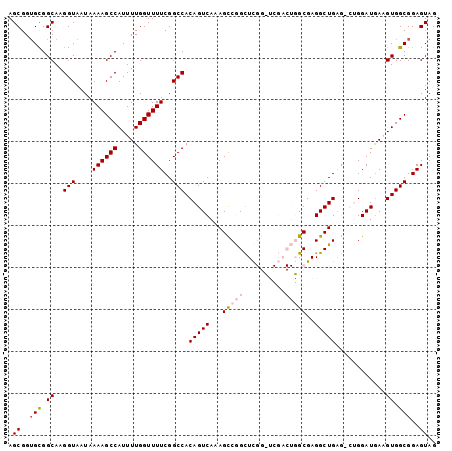
| Location | 9,722,345 – 9,722,454 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -29.80 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9722345 109 + 20766785 GUUUUCGGCCACAGUCAAAGCCGGCUCGGCUCGACUGGCGAGGCUGAAUCUGGAUAAAGUGGCGGAGUAGGAGGUGGCUGGUGGGACCUGGAGGUGGGUGUGGCCACAG ......(((((((.((...((((((.((.(((.(((..((..(((..((....))..)))..)).)))..))).))))))))...(((....))))).))))))).... ( -40.80) >DroSec_CAF1 40096 107 + 1 GUUUUCGGCCACAGUCAAAGCCGGCUCGGUUCGACUGGCGAGGCUGAGGCUGGAUGAAGUGGCGGAGUAGGAGGUGGCUGGUGGGACCUGGAGGUGGGUGUGGCC--AG ...(((.(((((..(((...(((((((((((((.....))).))))).))))).))).))))).)))..........(((((.(.((((......)))).).)))--)) ( -43.60) >DroSim_CAF1 31372 107 + 1 GUUUUCGGCCACAGUCAAAGCCGGCUCGGCUCGACUGGCGAGGCUGAGGCUGGAUGAAGUGGCGGAGUAGGAGGUGGCUGGUGGGACCUGGAGGUGGGUGUGGCC--AG ...(((.(((((..(((...(((((((((((((.....))).))))).))))).))).))))).)))..........(((((.(.((((......)))).).)))--)) ( -46.00) >DroEre_CAF1 32995 92 + 1 GUUUUCGGCCACAGUCGAAGCCAGCUC----------GCGAGGCUGGG-AAGGAUGAUGUGGCGGAGUAGGAGG---CUGGG-GGGCCUGGAGGUGGGUGUGGCC--AG ...(((.(((((((((....(((((.(----------....)))))).-...)))..)))))).))).....((---((...-(.((((......)))).)))))--.. ( -35.10) >consensus GUUUUCGGCCACAGUCAAAGCCGGCUCGGCUCGACUGGCGAGGCUGAGGCUGGAUGAAGUGGCGGAGUAGGAGGUGGCUGGUGGGACCUGGAGGUGGGUGUGGCC__AG ...(((.(((((.(((..((((.(((.(......).)))..)))).......)))...))))).)))............(((.(.((((......)))).).))).... (-29.80 = -30.18 + 0.38)

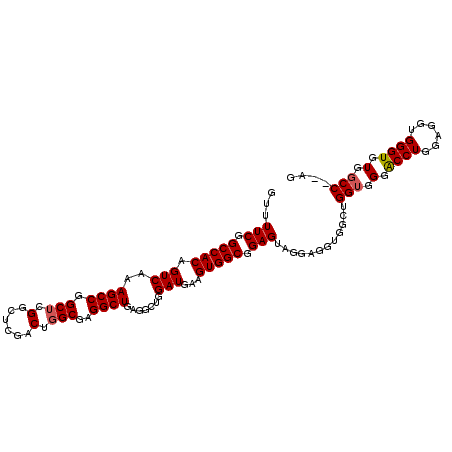
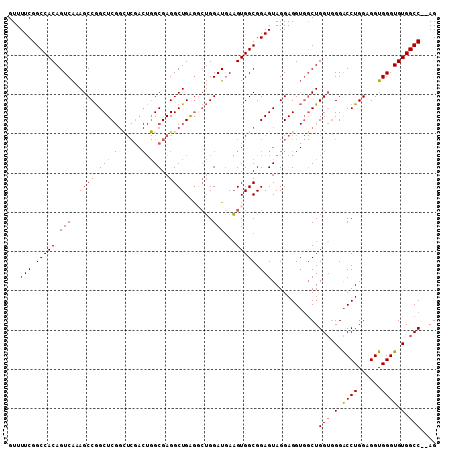
| Location | 9,722,345 – 9,722,454 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9722345 109 - 20766785 CUGUGGCCACACCCACCUCCAGGUCCCACCAGCCACCUCCUACUCCGCCACUUUAUCCAGAUUCAGCCUCGCCAGUCGAGCCGAGCCGGCUUUGACUGUGGCCGAAAAC ..(((((.......(((....))).......))))).........................(((.(((....(((((((((((...)))).))))))).))).)))... ( -30.64) >DroSec_CAF1 40096 107 - 1 CU--GGCCACACCCACCUCCAGGUCCCACCAGCCACCUCCUACUCCGCCACUUCAUCCAGCCUCAGCCUCGCCAGUCGAACCGAGCCGGCUUUGACUGUGGCCGAAAAC .(--(((.......(((....))).......))))........((.(((((.(((...((((...((.(((..........))))).)))).)))..))))).)).... ( -26.14) >DroSim_CAF1 31372 107 - 1 CU--GGCCACACCCACCUCCAGGUCCCACCAGCCACCUCCUACUCCGCCACUUCAUCCAGCCUCAGCCUCGCCAGUCGAGCCGAGCCGGCUUUGACUGUGGCCGAAAAC .(--(((.......(((....))).......))))........((.(((((........((....))......((((((((((...)))).))))))))))).)).... ( -28.34) >DroEre_CAF1 32995 92 - 1 CU--GGCCACACCCACCUCCAGGCCC-CCCAG---CCUCCUACUCCGCCACAUCAUCCUU-CCCAGCCUCGC----------GAGCUGGCUUCGACUGUGGCCGAAAAC .(--(((((((..(......((((..-....)---)))......................-.((((((....----------).)))))....)..))))))))..... ( -25.30) >consensus CU__GGCCACACCCACCUCCAGGUCCCACCAGCCACCUCCUACUCCGCCACUUCAUCCAGCCUCAGCCUCGCCAGUCGAGCCGAGCCGGCUUUGACUGUGGCCGAAAAC ....(((((((..(.......((.....))..................................((((..((..(......)..)).))))..)..)))))))...... (-19.51 = -19.57 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:19 2006