
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,720,810 – 9,720,920 |
| Length | 110 |
| Max. P | 0.560462 |

| Location | 9,720,810 – 9,720,920 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.57 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -23.71 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
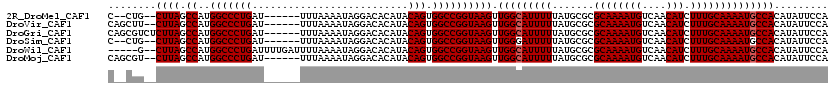
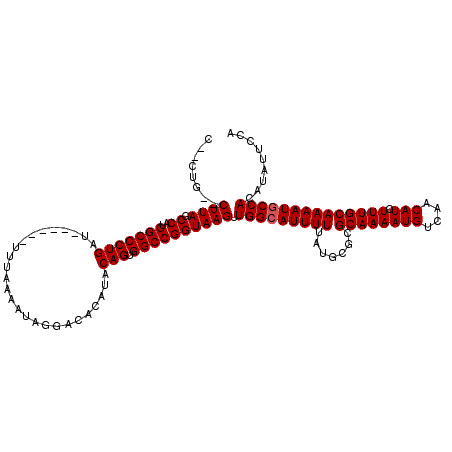
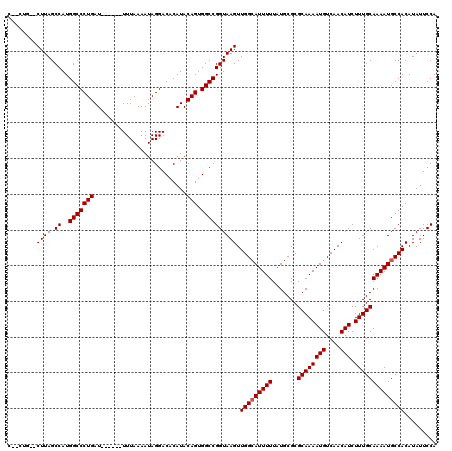
>2R_DroMel_CAF1 9720810 110 + 20766785 C--CUG--CUUAGCCAUGGCCCUGAU------UUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGCAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA .--..(--((((.((..(((((((..------..................))).)))))))))))(((((((((.......((((((((....))).))))))))))))))......... ( -26.76) >DroVir_CAF1 38128 112 + 1 CAGCUU--CUUAGCCAUGGCCCUGAU------UUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGCAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA ..(((.--...)))...(((((((..------..................))).))))((.....(((((((((.......((((((((....))).))))))))))))))......)). ( -26.66) >DroGri_CAF1 39287 114 + 1 CAGCGUCUCUUAGCCAUGGCCCUGAU------UUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGCAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA ..((........))...(((((((..------..................))).))))((.....(((((((((.......((((((((....))).))))))))))))))......)). ( -25.96) >DroSim_CAF1 29898 110 + 1 C--CUG--CUUAGCCAUGGCCCUGAU------UUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGGAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA .--(.(--((((.((..(((((((..------..................))).))))))))))).)(((....((((.(.(((...((.(((.....)))))...)))).)))).))). ( -23.65) >DroWil_CAF1 32125 113 + 1 -----G--CUUAGCCAUGGCCCUGAUUUUGAUUUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGCAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA -----(--((((.((..(((((((((((((....)))))).(....)...))).)))))))))))(((((((((.......((((((((....))).))))))))))))))......... ( -28.51) >DroMoj_CAF1 38880 112 + 1 CAGCGU--CUUAGCCAUGGCCCUGAU------UUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGCAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA ..((..--....))...(((((((..------..................))).))))((.....(((((((((.......((((((((....))).))))))))))))))......)). ( -26.16) >consensus C__CUG__CUUAGCCAUGGCCCUGAU______UUUAAAAUAGGACACAUACAGUGGCCGGUAAGUUGGCAUUUUUAUGCGCGCAAAAUGUCAACAUCUUUGCAAAAUGCCACAUAUUCCA ........((((.((..(((((((..........................))).)))))))))).(((((((((.......((((((((....))).))))))))))))))......... (-23.71 = -23.88 + 0.17)
| Location | 9,720,810 – 9,720,920 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.57 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.51 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
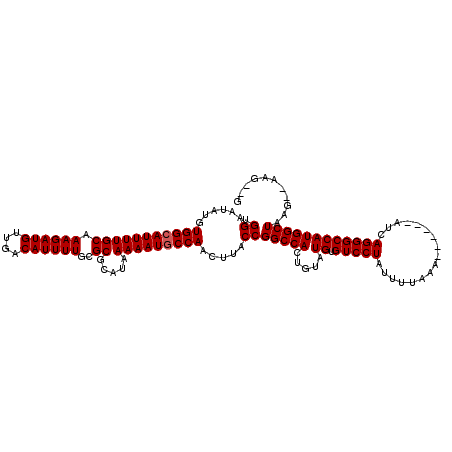
>2R_DroMel_CAF1 9720810 110 - 20766785 UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUGCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAA------AUCAGGGCCAUGGCUAAG--CAG--G .((......(((((((((((.((((((....))))))..)).....))))))))).....))(((((.....((.((((((((....)------)).))))))))))))...--...--. ( -28.40) >DroVir_CAF1 38128 112 - 1 UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUGCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAA------AUCAGGGCCAUGGCUAAG--AAGCUG .((......(((((((((((.((((((....))))))..)).....))))))))).....))((((.(((....((........))..------..)))))))..((((...--.)))). ( -30.70) >DroGri_CAF1 39287 114 - 1 UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUGCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAA------AUCAGGGCCAUGGCUAAGAGACGCUG .((......(((((((((((.((((((....))))))..)).....))))))))).....))((((.(((....((........))..------..)))))))..(((........))). ( -29.50) >DroSim_CAF1 29898 110 - 1 UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUCCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAA------AUCAGGGCCAUGGCUAAG--CAG--G .....(((((((((...(((((.....))).))...))).))))))......((..((((((((((.(((....((........))..------..)))))))..)).))))--..)--) ( -26.10) >DroWil_CAF1 32125 113 - 1 UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUGCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAAAUCAAAAUCAGGGCCAUGGCUAAG--C----- .((......(((((((((((.((((((....))))))..)).....))))))))).....))(((((.....((.((((((((((......))))).))))))))))))...--.----- ( -30.40) >DroMoj_CAF1 38880 112 - 1 UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUGCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAA------AUCAGGGCCAUGGCUAAG--ACGCUG .((......(((((((((((.((((((....))))))..)).....))))))))).....))((((.(((....((........))..------..)))))))..(((....--..))). ( -29.70) >consensus UGGAAUAUGUGGCAUUUUGCAAAGAUGUUGACAUUUUGCGCGCAUAAAAAUGCCAACUUACCGGCCACUGUAUGUGUCCUAUUUUAAA______AUCAGGGCCAUGGCUAAG__AAG__G .((......(((((((((((.((((((....))))))..)).....))))))))).....))(((((.....((.(((((.................))))))))))))........... (-26.35 = -26.51 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:16 2006