
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
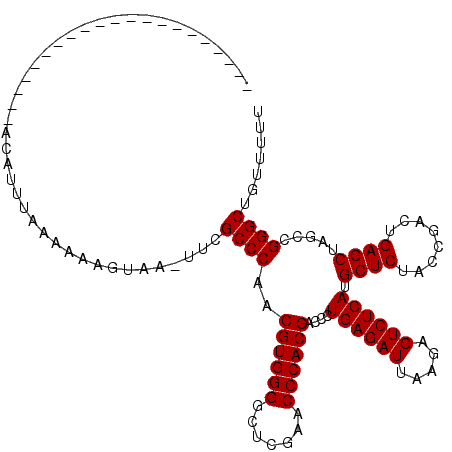
| Location | 1,678,865 – 1,678,963 |
| Length | 98 |
| Max. P | 0.988294 |

| Location | 1,678,865 – 1,678,963 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -31.17 |
| Energy contribution | -31.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
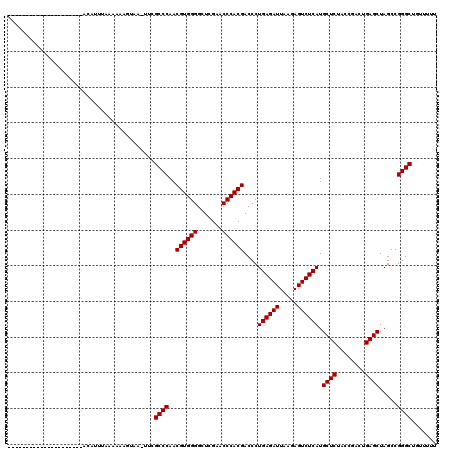
>2R_DroMel_CAF1 1678865 98 + 20766785 ---------------------ACAUUUAAAAAUAUAA-GCCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUUUUU ---------------------..............((-((.((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).)))).. ( -32.90) >DroPse_CAF1 94442 90 + 1 ---------------------------AAAAAUUUAUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAU---UU ---------------------------..............((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))..---.. ( -31.50) >DroSec_CAF1 63791 101 + 1 ------------------GUUACAUUUAAAAAAGUAA-UUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGCUUUU ------------------...........(((((((.-...((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))))))))) ( -33.50) >DroSim_CAF1 60960 101 + 1 ------------------GUUACAUUUAAAAAAGUAA-UUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUUUUC ------------------(((((..........))))-)..((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....... ( -31.40) >DroEre_CAF1 69597 119 + 1 CAAAAAGCCCUAUGCAAUAUUACAUCUAAAAAAAUAU-GUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUGCUG .....(((((((((.......((((..........))-)).......)))))))))......((((.(((((((((.....)))))).((((........)))).....))).))))... ( -33.14) >DroPer_CAF1 88354 90 + 1 ---------------------------AAAAAUGUACUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAU---UU ---------------------------..............((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))..---.. ( -31.50) >consensus _____________________ACAUUUAAAAAAGUAA_UUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUUUUU .........................................((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....... (-31.17 = -31.17 + 0.00)

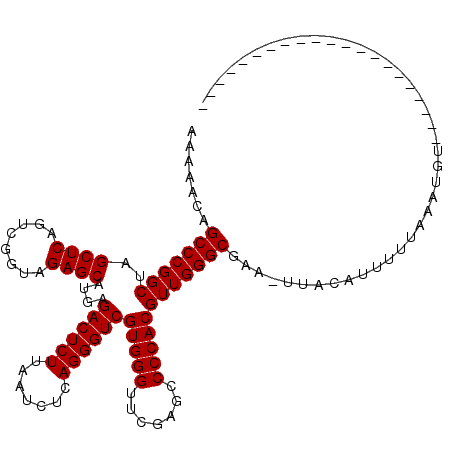
| Location | 1,678,865 – 1,678,963 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -32.83 |
| Energy contribution | -32.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
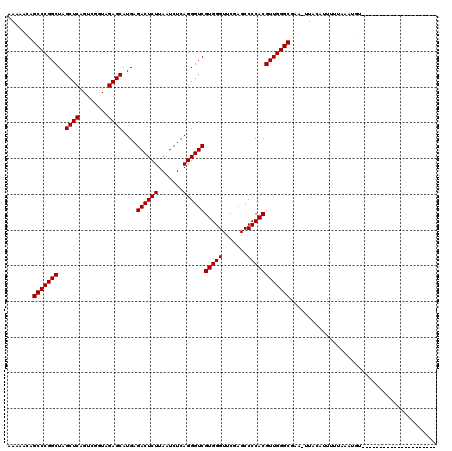
>2R_DroMel_CAF1 1678865 98 - 20766785 AAAAACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGGC-UUAUAUUUUUAAAUGU--------------------- .......(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))...-................--------------------- ( -32.90) >DroPse_CAF1 94442 90 - 1 AA---AUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAUAAAUUUUU--------------------------- ..---.((((((((..((((........))))....((((((.......))))))(((((.......))))))))))))).............--------------------------- ( -33.00) >DroSec_CAF1 63791 101 - 1 AAAAGCAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAA-UUACUUUUUUAAAUGUAAC------------------ .......(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))...-((((..........)))).------------------ ( -33.10) >DroSim_CAF1 60960 101 - 1 GAAAACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAA-UUACUUUUUUAAAUGUAAC------------------ .......(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))...-((((..........)))).------------------ ( -33.10) >DroEre_CAF1 69597 119 - 1 CAGCACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAC-AUAUUUUUUUAGAUGUAAUAUUGCAUAGGGCUUUUUG .(((...(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))...-............(((((....)))))...)))..... ( -37.40) >DroPer_CAF1 88354 90 - 1 AA---AUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAGUACAUUUUU--------------------------- ..---.((((((((..((((........))))....((((((.......))))))(((((.......))))))))))))).............--------------------------- ( -33.00) >consensus AAAAACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAA_UUACAUUUUUAAAUGU_____________________ .......(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))......................................... (-32.83 = -32.83 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:24 2006