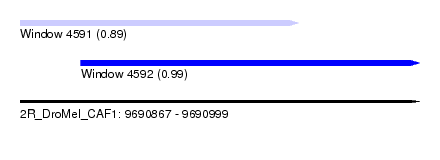
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,690,867 – 9,690,999 |
| Length | 132 |
| Max. P | 0.989420 |

| Location | 9,690,867 – 9,690,959 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.10 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -21.97 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9690867 92 + 20766785 CGUUGUCAUGCGAAUAUACGAAUAUGUACGAAUGAACGUACGUAUGUAUGUUUUUAUGGGGGGGAUGGAGCGAGUGUAUUAGAAUUCUUUUA (.((.(((((.((((((((..(((((((((......))))))))))))))))).))))).)).).......((((........))))..... ( -23.60) >DroSec_CAF1 7263 91 + 1 CGUUGUCAUGCGAAUAUACGAAUAUGUACGAAUGAACGUACGUAUGUAUGUUAAUA-GGGGGGGAUGGAGCGAGUGUGUUAGAAUUCUUUUA (((......)))(((((((..(((((((((......))))))))))))))))....-.((((((((..((((....))))...)))))))). ( -21.40) >DroSim_CAF1 5411 92 + 1 CGUUGUCAUGCGAAUAUACGAAUAUGUACGAAUGAACGUACGUAUGUAUGUUUAUAUGGGGGGGAUGGAGCGAGUGUGUUAGAAUUCUUUUA ......((((..(((((((..(((((((((......))))))))))))))))..))))((((((((..((((....))))...)))))))). ( -25.30) >consensus CGUUGUCAUGCGAAUAUACGAAUAUGUACGAAUGAACGUACGUAUGUAUGUUUAUAUGGGGGGGAUGGAGCGAGUGUGUUAGAAUUCUUUUA (.((.(((((..(((((((..(((((((((......))))))))))))))))..))))).)).).......((((........))))..... (-21.97 = -22.30 + 0.33)

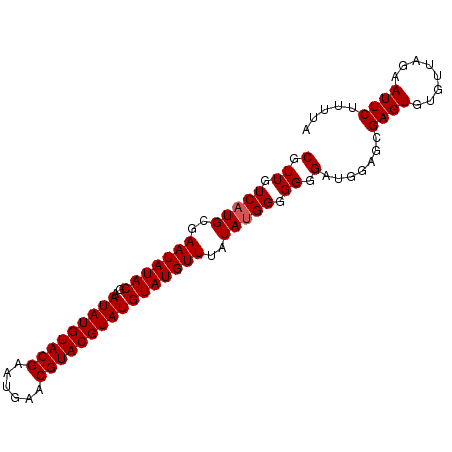
| Location | 9,690,887 – 9,690,999 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -24.16 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9690887 112 + 20766785 AAUAUGUACGAAUGAACGUACGUAUGUAUGUUUUUAUGGGGGGGAUGGAGCGAGUGUAUUAGAAUUCUUUUAGCAUACAUCCAUACGGAUUUUUUUCGUUUCCAUUUCAUUC .(((((((((......)))))))))................(..(((((((((((((.(((((.....))))).))))((((....)))).....))).))))))..).... ( -29.50) >DroSec_CAF1 7283 111 + 1 AAUAUGUACGAAUGAACGUACGUAUGUAUGUUAAUA-GGGGGGGAUGGAGCGAGUGUGUUAGAAUUCUUUUACCAUACAUCCAUACGGAUUUUUUUCGUUUCCAUUUCAUUC .(((((((((......)))))))))...........-....(..((((((((((((((.((((.....)))).)))))((((....)))).....))).))))))..).... ( -31.60) >DroSim_CAF1 5431 112 + 1 AAUAUGUACGAAUGAACGUACGUAUGUAUGUUUAUAUGGGGGGGAUGGAGCGAGUGUGUUAGAAUUCUUUUACCAUACAUCCAUACGGAUUUUUUUCGUUUCCAUUUCAUUC ........((.((((((((((....)))))))))).))...(..((((((((((((((.((((.....)))).)))))((((....)))).....))).))))))..).... ( -31.90) >DroEre_CAF1 5344 90 + 1 AA----------------------UGUAUAUUUAUAUGGGGGGAAUGGAGCGAGUGUGUUAGAAUUCUUUUACCAUACAUCCAUACGGGUUUUUUUCGUUUCCAUUUCAUUC .(----------------------((((....)))))....(((((((((((((((((.((((.....)))).)))))((((....)))).....))).))))))))).... ( -22.50) >DroYak_CAF1 5374 102 + 1 AAUAUGUACGU----------UUAUGUAUGUUUAUAUGGGGGGAAUGGAGCGAGUGUGUUAGAAUUCUUUUACCAUACAUCCAUAGGGGUUUUUUUCGUUUCCAUUUCAUUC .(((((.((((----------......)))).)))))....(((((((((((((((((.((((.....)))).)))))((((....)))).....))).))))))))).... ( -25.30) >consensus AAUAUGUACGAAUGAACGUACGUAUGUAUGUUUAUAUGGGGGGGAUGGAGCGAGUGUGUUAGAAUUCUUUUACCAUACAUCCAUACGGAUUUUUUUCGUUUCCAUUUCAUUC .(((((((((......)))))))))................(((((((((((((((((.((((.....)))).)))))((((....)))).....))).))))))))).... (-24.16 = -25.48 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:08 2006