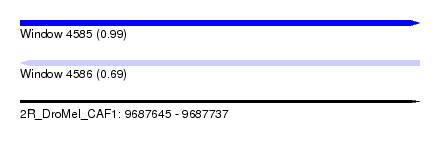
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,687,645 – 9,687,737 |
| Length | 92 |
| Max. P | 0.988756 |

| Location | 9,687,645 – 9,687,737 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -19.34 |
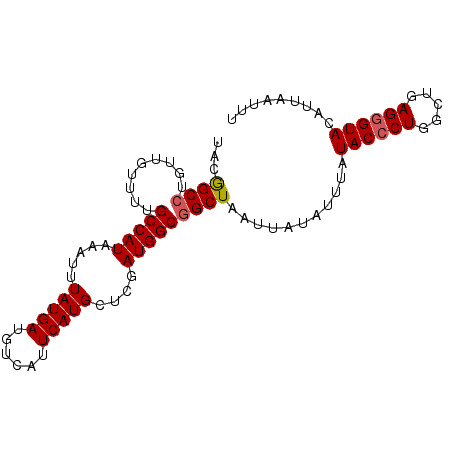
| Consensus MFE | -16.44 |
| Energy contribution | -16.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9687645 92 + 20766785 AAAUUAAAGUACCCUCAGCCAGGGUAUAAAUAUAAUUAGCCGCCAUCGAGCAUGAAUGACAUCAUAAAUUUAUGGCAAAACAACAGGCCGUA ........(((((((.....)))))))...........((((((((.((..((((......))))...)).))))).........))).... ( -18.90) >DroSec_CAF1 4039 92 + 1 AAAUUAAUAUACCCUCAGCCAGGGUAUAAAUAUAAUUAGCCGCCAUCGGGCAUGAAUGACAUCAUAAAUUUAUGGCAAAACAACAGGCCGUA .......((((((((.....))))))))..........((((((....)))((((......))))........)))................ ( -20.90) >DroSim_CAF1 2191 92 + 1 AAAUUAAUGUACCCUCAGCCAGGGUAUAAAUAUAAUUAGCCGCCAUCGGGCAUGAAUGACAUCAUAAAUUUAUGGCAAAACAACAGGCCGUA .......((((((((.....))))))))..........((((((....)))((((......))))........)))................ ( -20.90) >DroEre_CAF1 2143 92 + 1 AAAAUAAUGUACACUCUGGUAGGGUAUAAAUAUAAUUAGCCGCCAUCGAGCAUGAAUGACAUCAUAAAUUUAUGGCAAAACAACAGGCUGUA .........((((..(((...((.((..........)).))(((((.((..((((......))))...)).))))).......)))..)))) ( -18.10) >DroYak_CAF1 2154 88 + 1 ----CAAAGUACCCUCUGCUAGGGUAUAAAUAUAAUUAGCCGCCAUCGAGCAUGAAUGACAUCAUAAAUUUAUGGCAAACCAACACGCCGUA ----....(((((((.....)))))))..............(((((.((..((((......))))...)).)))))................ ( -17.90) >consensus AAAUUAAUGUACCCUCAGCCAGGGUAUAAAUAUAAUUAGCCGCCAUCGAGCAUGAAUGACAUCAUAAAUUUAUGGCAAAACAACAGGCCGUA ........(((((((.....)))))))...........((((((((..(..((((......))))...)..))))).........))).... (-16.44 = -16.44 + -0.00)

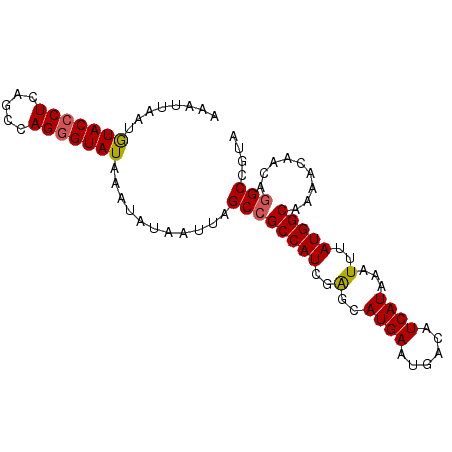
| Location | 9,687,645 – 9,687,737 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9687645 92 - 20766785 UACGGCCUGUUGUUUUGCCAUAAAUUUAUGAUGUCAUUCAUGCUCGAUGGCGGCUAAUUAUAUUUAUACCCUGGCUGAGGGUACUUUAAUUU ...((((.........(((((.....(((((......)))))....)))))))))...........((((((.....))))))......... ( -21.10) >DroSec_CAF1 4039 92 - 1 UACGGCCUGUUGUUUUGCCAUAAAUUUAUGAUGUCAUUCAUGCCCGAUGGCGGCUAAUUAUAUUUAUACCCUGGCUGAGGGUAUAUUAAUUU ...((((.........(((((.....(((((......)))))....)))))))))(((((....((((((((.....)))))))).))))). ( -23.90) >DroSim_CAF1 2191 92 - 1 UACGGCCUGUUGUUUUGCCAUAAAUUUAUGAUGUCAUUCAUGCCCGAUGGCGGCUAAUUAUAUUUAUACCCUGGCUGAGGGUACAUUAAUUU ...((((.........(((((.....(((((......)))))....)))))))))...........((((((.....))))))......... ( -21.10) >DroEre_CAF1 2143 92 - 1 UACAGCCUGUUGUUUUGCCAUAAAUUUAUGAUGUCAUUCAUGCUCGAUGGCGGCUAAUUAUAUUUAUACCCUACCAGAGUGUACAUUAUUUU ...((((.........(((((.....(((((......)))))....))))))))).........(((((.(.....).)))))......... ( -15.40) >DroYak_CAF1 2154 88 - 1 UACGGCGUGUUGGUUUGCCAUAAAUUUAUGAUGUCAUUCAUGCUCGAUGGCGGCUAAUUAUAUUUAUACCCUAGCAGAGGGUACUUUG---- ........(((((((.(((((.....(((((......)))))....))))))))))))........((((((.....)))))).....---- ( -22.30) >consensus UACGGCCUGUUGUUUUGCCAUAAAUUUAUGAUGUCAUUCAUGCUCGAUGGCGGCUAAUUAUAUUUAUACCCUGGCUGAGGGUACAUUAAUUU ...((((.........(((((.....(((((......)))))....)))))))))...........((((((.....))))))......... (-18.76 = -19.00 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:01 2006