| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
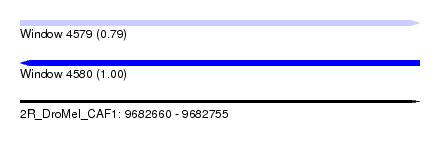
| Location | 9,682,660 – 9,682,755 |
| Length | 95 |
| Max. P | 0.999924 |

| Location | 9,682,660 – 9,682,755 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.05 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
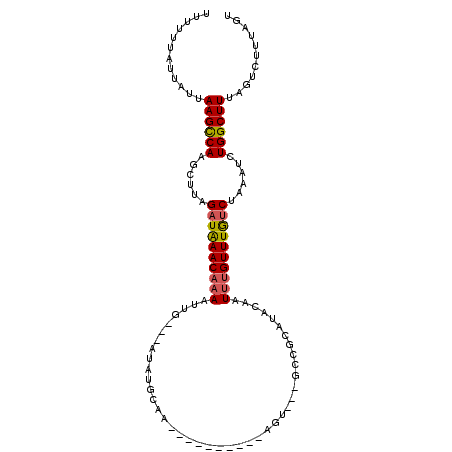
>2R_DroMel_CAF1 9682660 95 + 20766785 CUUUUUAUUAUUAAGCCAAGCUUUGAUAAACAAAUUUG---AUAUGCA-----------AGUGCCGCCGCAUACAAUUUGUUUGUCUAAAUCUGGCUUUAGUCUUUAGU ............((((((.(.(((((((((((((((.(---.(((((.-----------.........)))))))))))))))))).))).)))))))........... ( -23.20) >DroSec_CAF1 5132 103 + 1 UUUUUUAUUAUUAAGCCAAGCUUAGAUAAACAAAAUUG---AUGUGCAACAGAAUGUAAAGU---GCCGCAUACAAUUUGUUUAUCCAAAUCUGGCUUUAGUCUUUAGU ............((((((......((((((((((.(((---(((((((((.....))....)---).))))).)))))))))))))......))))))........... ( -22.40) >DroSim_CAF1 5153 103 + 1 UUUUUUAUUUUUAAGCCAAGCUUAGAUAAACACAAUUG---AUAUGCAGCCGAAUGUAAAGU---GCCUCAUACAAUUUGUUUGUCCAAAUCUGGCUUUAGUCUUUAGU ............((((((......((((((((.(((((---.((((..(((.........).---))..)))))))))))))))))......))))))........... ( -21.70) >DroEre_CAF1 5250 81 + 1 UUUUUUAUUUUUAAGUCAAGUUCAGAUGAACCAAAUUU---AUAUGCAA-------------------------CUUUUGUUUGUCUAAAUCUGGCUUAAGCUUGUAGC .........(((((((((..((.(((..(((.(((.((---......))-------------------------.))).)))..))).))..)))))))))........ ( -16.00) >DroYak_CAF1 5210 96 + 1 UUUUUUAUUGUUAAGCCAAGUUCAGAUAAACAAAAUUGAAUGUAUGUCG----------GUU---AUCGCUUAUAAUUUGUUUGCCUAAAUCUGGCUUUAGUCUGUAGC ....((((.(..((((((..((.((.((((((((..(((.((.(((...----------..)---)))).)))...)))))))).)).))..))))))....).)))). ( -16.00) >consensus UUUUUUAUUAUUAAGCCAAGCUUAGAUAAACAAAAUUG___AUAUGCAA__________AGU___GCCGCAUACAAUUUGUUUGUCUAAAUCUGGCUUUAGUCUUUAGU ............((((((......((((((((((..........................................))))))))))......))))))........... (-10.85 = -10.97 + 0.12)


| Location | 9,682,660 – 9,682,755 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.05 |
| Mean single sequence MFE | -23.31 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.11 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
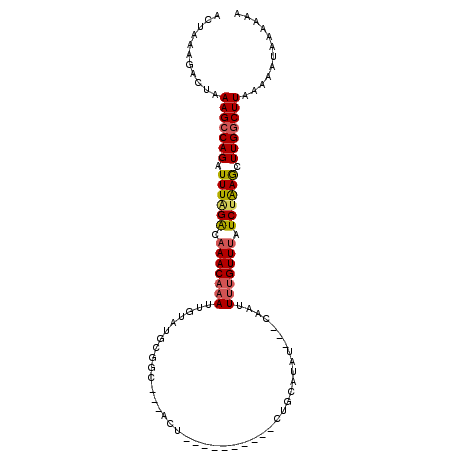
>2R_DroMel_CAF1 9682660 95 - 20766785 ACUAAAGACUAAAGCCAGAUUUAGACAAACAAAUUGUAUGCGGCGGCACU-----------UGCAUAU---CAAAUUUGUUUAUCAAAGCUUGGCUUAAUAAUAAAAAG ...........(((((((.(((.((.(((((((((((((((((......)-----------)))))))---..))))))))).)).))).)))))))............ ( -24.40) >DroSec_CAF1 5132 103 - 1 ACUAAAGACUAAAGCCAGAUUUGGAUAAACAAAUUGUAUGCGGC---ACUUUACAUUCUGUUGCACAU---CAAUUUUGUUUAUCUAAGCUUGGCUUAAUAAUAAAAAA ...........(((((((.(((((((((((((((((..((((((---(..........)))))))...---))).)))))))))))))).)))))))............ ( -32.20) >DroSim_CAF1 5153 103 - 1 ACUAAAGACUAAAGCCAGAUUUGGACAAACAAAUUGUAUGAGGC---ACUUUACAUUCGGCUGCAUAU---CAAUUGUGUUUAUCUAAGCUUGGCUUAAAAAUAAAAAA ...........(((((((.((((((.((((((((((((((.(((---............))).)))).---))))).))))).)))))).)))))))............ ( -26.70) >DroEre_CAF1 5250 81 - 1 GCUACAAGCUUAAGCCAGAUUUAGACAAACAAAAG-------------------------UUGCAUAU---AAAUUUGGUUCAUCUGAACUUGACUUAAAAAUAAAAAA ....((((.((((((((((((((....(((....)-------------------------)).....)---))))))))))....))).))))................ ( -14.80) >DroYak_CAF1 5210 96 - 1 GCUACAGACUAAAGCCAGAUUUAGGCAAACAAAUUAUAAGCGAU---AAC----------CGACAUACAUUCAAUUUUGUUUAUCUGAACUUGGCUUAACAAUAAAAAA ...........(((((((.(((((..(((((((...........---...----------...............)))))))..))))).)))))))............ ( -18.45) >consensus ACUAAAGACUAAAGCCAGAUUUAGACAAACAAAUUGUAUGCGGC___ACU__________CUGCAUAU___CAAUUUUGUUUAUCUAAGCUUGGCUUAAAAAUAAAAAA ...........(((((((.((((((.(((((((..........................................))))))).)))))).)))))))............ (-13.91 = -14.11 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:55 2006