
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,676,230 – 9,676,334 |
| Length | 104 |
| Max. P | 0.763413 |

| Location | 9,676,230 – 9,676,334 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -22.73 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
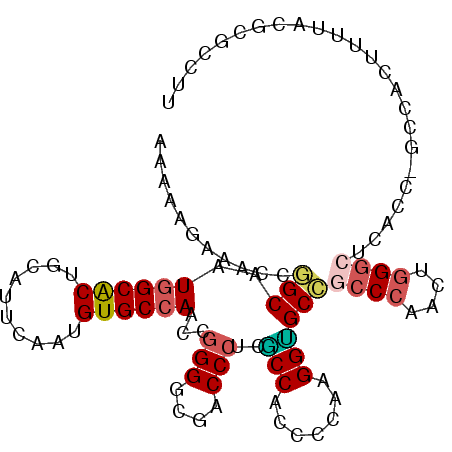
>2R_DroMel_CAF1 9676230 104 + 20766785 AAGAAUAAACCUGCAAUGGCGCUGCAUUCAAUGUGCCAACCGGGGCGUCCCUCGCCACCCCAGGGUGCAGCCCACGUGGGCUCACU-GCCACUUUUACGCGCCUU .................(((((.((((.....)))).....((((((.....)))).......((((.(((((....)))))))))-.))........))))).. ( -36.30) >DroPse_CAF1 2484 104 + 1 AAAAAGAAUCCGGCAAUGGCACUGCAUUCAAUGUGCCAGCCGGGGCGACCCUCUCCACCCCAAGGAGCCGCCCAACUGGGCUCAUCAGCCUCCUUCU-UGGCCUU ...(((((.(((((..((((((..........)))))))))))(((......((((.......))))..((((....))))......)))...))))-)...... ( -38.30) >DroEre_CAF1 2451 104 + 1 AAGAAAAGACCUGCAAUGGCACUGCAUUCGAUGUGCCAACCGGGGCGACCCUCGCCACCCCAAGGUGCAGCCCAGGUGGGCUCACC-GCUACUUUUACGCGCCUU ...(((((....((..((((((..........))))))...((((((.....)))).))....((((.(((((....)))))))))-))..)))))......... ( -38.60) >DroYak_CAF1 2451 104 + 1 AAGAAAAGACCAGCAAUGGCGCUACAUUCGAUGUGCCAACCGGGGCGUCCCUCACCACCCCAAGGUGCCUCCCAGGUGGGCUCACU-GCCACUUUUACGCGCCUU ................((((((..........))))))...((((.(........).))))(((((((.....((((((.(.....-)))))))....))))))) ( -32.60) >DroAna_CAF1 2420 104 + 1 AAAAAGAGGCCGGCAAUAGCACUACACUCGAUGUGCCAGCCAGGACGUCCCUCGCCGCCCCACGGCGCCUGCCAACUUGGCUCCUU-UUGGGUUUUGCGCGCCUU .....(((((.(((....((((..........))))..((.(((.....))).)).))).....((((..(((.....)))(((..-..)))....))))))))) ( -33.00) >DroPer_CAF1 2484 104 + 1 AAAAAGAAUCCGGCAAUGGCACUGCAUUCAAUGUGCCAGCCGGGGCGACCCUCUCCACCCCAAGGAGCCGCCCAACUGGGCUCAUCAGCCUCCUUCU-UGGCCUU ...(((((.(((((..((((((..........)))))))))))(((......((((.......))))..((((....))))......)))...))))-)...... ( -38.30) >consensus AAAAAGAAACCGGCAAUGGCACUGCAUUCAAUGUGCCAACCGGGGCGACCCUCGCCACCCCAAGGUGCCGCCCAACUGGGCUCACC_GCCACUUUUACGCGCCUU ...........(((..((((((..........))))))...(((....)))..(((.......))))))((((....))))........................ (-22.73 = -22.73 + 0.00)

| Location | 9,676,230 – 9,676,334 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -42.58 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
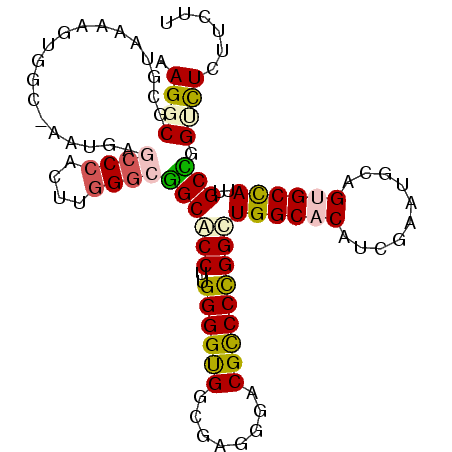
>2R_DroMel_CAF1 9676230 104 - 20766785 AAGGCGCGUAAAAGUGGC-AGUGAGCCCACGUGGGCUGCACCCUGGGGUGGCGAGGGACGCCCCGGUUGGCACAUUGAAUGCAGCGCCAUUGCAGGUUUAUUCUU ..((((((((..((((((-.((((((((....))))).))).((((((((........))))))))...)).))))...))).)))))................. ( -45.20) >DroPse_CAF1 2484 104 - 1 AAGGCCA-AGAAGGAGGCUGAUGAGCCCAGUUGGGCGGCUCCUUGGGGUGGAGAGGGUCGCCCCGGCUGGCACAUUGAAUGCAGUGCCAUUGCCGGAUUCUUUUU ......(-((((((((((......((((....))))((((((((......))).))))))))(((((((((((..........))))))..))))).)))))))) ( -43.10) >DroEre_CAF1 2451 104 - 1 AAGGCGCGUAAAAGUAGC-GGUGAGCCCACCUGGGCUGCACCUUGGGGUGGCGAGGGUCGCCCCGGUUGGCACAUCGAAUGCAGUGCCAUUGCAGGUCUUUUCUU (((((((......)).((-(((((((((....))))).))))((((((((((....)))))))))).((((((..........))))))..))..)))))..... ( -45.80) >DroYak_CAF1 2451 104 - 1 AAGGCGCGUAAAAGUGGC-AGUGAGCCCACCUGGGAGGCACCUUGGGGUGGUGAGGGACGCCCCGGUUGGCACAUCGAAUGUAGCGCCAUUGCUGGUCUUUUCUU ..(((((.((...((..(-.(((.(((.(((.((......))..((((((........))))))))).))).))).).)).)))))))................. ( -36.10) >DroAna_CAF1 2420 104 - 1 AAGGCGCGCAAAACCCAA-AAGGAGCCAAGUUGGCAGGCGCCGUGGGGCGGCGAGGGACGUCCUGGCUGGCACAUCGAGUGUAGUGCUAUUGCCGGCCUCUUUUU ..(((((.......((..-..)).(((.....)))..)))))..(((((((((((((....)))(((..((((.....))))...))).))))).)))))..... ( -42.20) >DroPer_CAF1 2484 104 - 1 AAGGCCA-AGAAGGAGGCUGAUGAGCCCAGUUGGGCGGCUCCUUGGGGUGGAGAGGGUCGCCCCGGCUGGCACAUUGAAUGCAGUGCCAUUGCCGGAUUCUUUUU ......(-((((((((((......((((....))))((((((((......))).))))))))(((((((((((..........))))))..))))).)))))))) ( -43.10) >consensus AAGGCGCGUAAAAGUGGC_AAUGAGCCCACUUGGGCGGCACCUUGGGGUGGCGAGGGACGCCCCGGCUGGCACAUCGAAUGCAGUGCCAUUGCCGGUCUCUUCUU .((((...................((((....))))((((((..((((((........)))))))))((((((..........))))))..))).))))...... (-28.16 = -28.25 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:47 2006