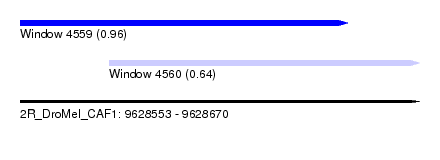
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,628,553 – 9,628,670 |
| Length | 117 |
| Max. P | 0.955281 |

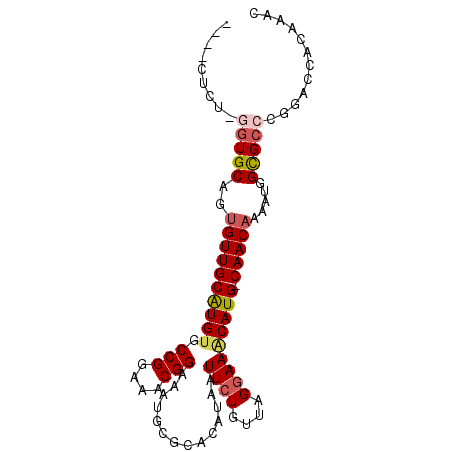
| Location | 9,628,553 – 9,628,649 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -15.36 |
| Energy contribution | -14.83 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9628553 96 + 20766785 GAAGA------CUCCGUUUUCCGC----GCUG-AC--UG----CUCUCGGUGCAUUG--UUGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAGCAUG-CAACAA--- (((((------(...)))))).((----((((-(.--..----...))))))).(((--((((((((.(((....)))..............((((....)))))))))-))))))--- ( -35.60) >DroVir_CAF1 31365 105 + 1 GAAU-------CGCCAUUUUGUUUGCGCUCUGCUC--GG----CUCUUGGUGCACUGUGUUGUGUGUACCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACGUG-GUACAGUCC ....-------.(((((.((((.(((((....(.(--(.----.(((.(((((((........))))))))))..)))....))))).))))(((.....)))...)))-))....... ( -31.50) >DroEre_CAF1 27643 96 + 1 GAAGA------CGCCAUUUUCCGC----GCUG-AC--GG----CACUCGGUGCAUUG--UUGCAUGUGCCGGAAACGGAAACGCGCACAUAAUUCUGUUAGGAAGCAUG-CAACAA--- (((((------.....))))).((----((((-(.--..----...))))))).(((--((((((((.(((....)))..............((((....)))))))))-))))))--- ( -33.60) >DroWil_CAF1 28894 83 + 1 GAAGACG---ACGCCAUUUUGU---------------------------GUGCAGCG--UUGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACAUG-CAACAA--- .......---..((....((((---------------------------((((.(((--((.......(((....))).)))))))))))))...((((....)))).)-).....--- ( -22.70) >DroYak_CAF1 37057 83 + 1 GAAAG------CGCCAUUUUCUGC------------------------GGUGCACUG--UUGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAGCAUG-CAACAA--- ....(------((((.........------------------------)))))..((--((((((((.(((....)))..............((((....)))))))))-))))).--- ( -30.40) >DroPer_CAF1 21618 109 + 1 GAAGAAGAAUAUGCCAUUUUGCUC----GCUG-AUGCUGCGCUCUCUUGGUGCAGUG--UUGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUGCCAACAA--- ..((((..((.((((((((.((.(----((.(-((((((((((.....)))))))))--))))).)).(((....)))))))).))).))..))))(((.((.......)))))..--- ( -37.30) >consensus GAAGA______CGCCAUUUUCCGC____GCUG_AC___G____CUCU_GGUGCACUG__UUGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUG_CAACAA___ .................................................((((........((((...(((....)))..)))))))).......((((....))))............ (-15.36 = -14.83 + -0.53)

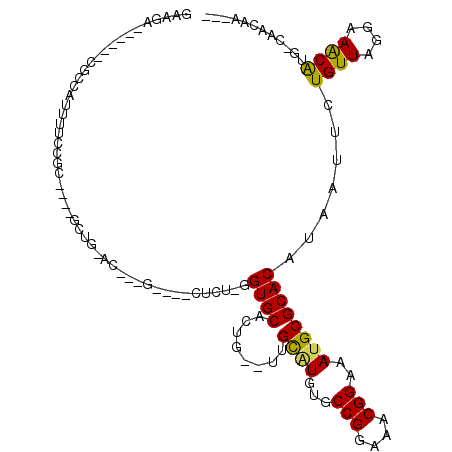
| Location | 9,628,579 – 9,628,670 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 85.05 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -22.27 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9628579 91 + 20766785 ----CUCUCGGUGCAUUGUUGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAGCAUG-CAACAAAAUGGCGCCCGGACCACAUCA ----.(((.(((((.(((((((((((.(((....)))..............((((....)))))))))-))))))....))))).)))........ ( -34.50) >DroPse_CAF1 26874 96 + 1 CGCUCUCUUGGUGCAGUGUUGCGUGGGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUGCCAACAAAAUGGCGCCCGCACCGCAGAC .....(((((((((.((((.((((...(((....)))..))))))))............(....)..((((......))))....)))))).))). ( -35.80) >DroEre_CAF1 27669 90 + 1 ----CACUCGGUGCAUUGUUGCAUGUGCCGGAAACGGAAACGCGCACAUAAUUCUGUUAGGAAGCAUG-CAACAAAAUGGCGC-CGGACCACAAAC ----...(((((((.(((((((((((.(((....)))..............((((....)))))))))-))))))....))))-)))......... ( -35.30) >DroWil_CAF1 28913 85 + 1 ----------GUGCAGCGUUGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACAUG-CAACAAAAUGGUGAAUGGACCACCAAC ----------.......(((((((((.(((....)))..............(((.....))).)))))-))))....(((((.......))))).. ( -26.30) >DroYak_CAF1 37075 86 + 1 ---------GGUGCACUGUUGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAGCAUG-CAACAAAAUGGCGCCCGGACCACAACC ---------(((((..((((((((((.(((....)))..............((((....)))))))))-))))).....)))))............ ( -30.50) >DroPer_CAF1 21652 96 + 1 CGCUCUCUUGGUGCAGUGUUGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUGCCAACAAAAUGGCGCCCGCACCGCAGAC .....(((((((((.((((.((((...(((....)))..))))))))............(....)..((((......))))....)))))).))). ( -35.80) >consensus ____CUCU_GGUGCAGUGUUGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUG_CAACAAAAUGGCGCCCGGACCACAAAC .........(((((..((((((((((.(((....)))..............((((....))))))))).))))).....)))))............ (-22.27 = -22.72 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:35 2006