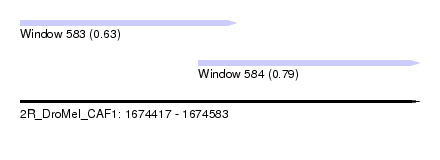
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,674,417 – 1,674,583 |
| Length | 166 |
| Max. P | 0.789850 |

| Location | 1,674,417 – 1,674,507 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
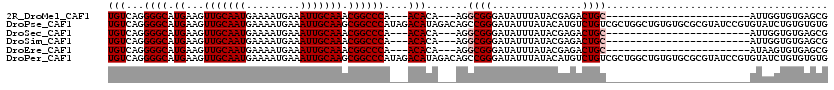
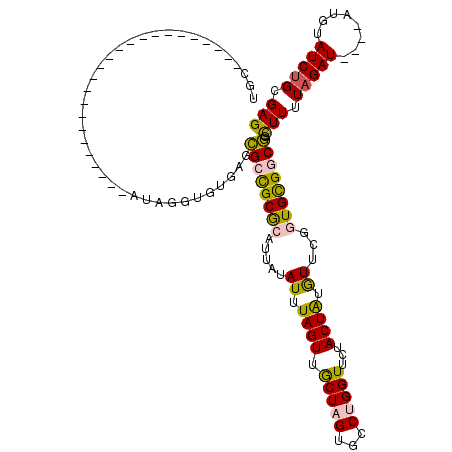
>2R_DroMel_CAF1 1674417 90 + 20766785 UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAACGGCCCA---ACACA---AGGCGGGAUAUUUAUACGAGACUGC------------------------AUUGGUGUGAGCG ......((((......(((((((.........)))))))...)))).---((((.---..((((...............))))------------------------....))))..... ( -23.66) >DroPse_CAF1 91572 120 + 1 UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAGCGGCCCAUAGACAUAGACAGCCGGGAUAUUUAUACAUGUCUGUCGCUGGCUGUGUGCGCGUAUCCGUGUAUCUGUGUGUG ......((((.((...(((((((.........))))))).)))))).((.((((((((((((((((((..((....))..)))).))))))))(((((((....))))))))))))).)) ( -43.40) >DroSec_CAF1 58429 90 + 1 UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAACGGCCCA---ACACA---AGGCGGGAUAUUUAUACGAGACUGC------------------------AUUGGUGUGAGCG ......((((......(((((((.........)))))))...)))).---((((.---..((((...............))))------------------------....))))..... ( -23.66) >DroSim_CAF1 55420 90 + 1 UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAACGGCCCA---ACACA---AGGCGGGAUAUUUAUACGAGACUGC------------------------AUUGGUGUGAGCG ......((((......(((((((.........)))))))...)))).---((((.---..((((...............))))------------------------....))))..... ( -23.66) >DroEre_CAF1 65833 90 + 1 UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAACGGCCCA---ACACA---AGGCGGGAUAUUUAUACGAGACUGC------------------------AUAAGUGUGAGCG ......((((......(((((((.........)))))))...)))).---.((((---..((((...............))))------------------------.....)))).... ( -22.96) >DroPer_CAF1 85460 120 + 1 UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAGCGGCCCAUAGACAUAGACAGCCGGGAUAUUUAUACAUGUCUGUCGCUGGCUGUGUGCGCGUAUCCGUGUAUCUGUGUGUG ......((((.((...(((((((.........))))))).)))))).((.((((((((((((((((((..((....))..)))).))))))))(((((((....))))))))))))).)) ( -43.40) >consensus UGUCAGGGGCAUGAAGUUGCAAUGAAAAUGAAAUUGCAAACGGCCCA___ACACA___AGGCGGGAUAUUUAUACGAGACUGC________________________AUAGGUGUGAGCG (((...((((.((...(((((((.........))))))).))))))....))).......((((...............))))..................................... (-15.73 = -16.06 + 0.33)
| Location | 1,674,491 – 1,674,583 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -18.57 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
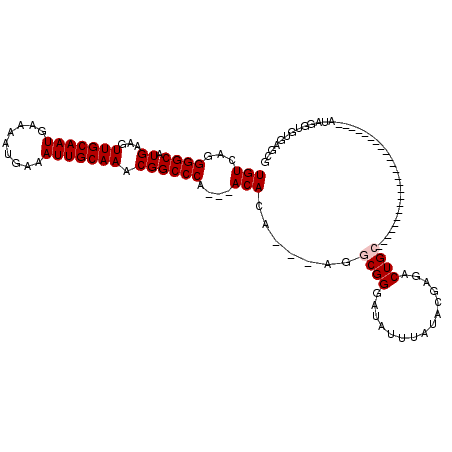
>2R_DroMel_CAF1 1674491 92 + 20766785 UGC------------------------AUUGGUGUGAGCGCCGCGCAUUAUAUUUAGUUGCUAGUGCCUGGUUCCACUAUGUUCGGUGUGGCGGCUUUUAGAU----AUGUAUCUGCGAG (((------------------------((......(((((((((((...((((...(..(((((...)))))..)...))))...))))))).))))......----)))))........ ( -26.60) >DroPse_CAF1 91652 116 + 1 UGUCGCUGGCUGUGUGCGCGUAUCCGUGUAUCUGUGUGUGGUGCACAUUAUAUUUAGUUACUUGUGUCUGGUUCUACUGCAUUCUAUGCGGCAGUUUCUAGAU----AUGUAUCUGCGAG ..((((((((((.(((((((....))))))).(((((.....))))).......))))))...((((((((..((.((((((...)))))).))...))))))----))......)))). ( -34.60) >DroSec_CAF1 58503 92 + 1 UGC------------------------AUUGGUGUGAGCGCCGCGCAUUAUAUUUAGUUGCUAGUGCCUGGUUCUACUAUGUUCGGUGUGGCGGCUUUUAGAU----AUGUAUCUGCGAG (((------------------------((......(((((((((((...((((.(((..(((((...))))).)))..))))...))))))).))))......----)))))........ ( -27.20) >DroSim_CAF1 55494 92 + 1 UGC------------------------AUUGGUGUGAGCGCCGCGCAUUAUAUUUAGUUGCUAGUGCCUGGUUCUACUAUGUUCGGUGUGGCGGCUUUUAGAU----AUGUAUCUGCGAG (((------------------------((......(((((((((((...((((.(((..(((((...))))).)))..))))...))))))).))))......----)))))........ ( -27.20) >DroEre_CAF1 65907 96 + 1 UGC------------------------AUAAGUGUGAGCGCCGCGCAUUAUAUUUAGUUGCUAGUGCCUGGUUCUACUAUGUUCUGUGCGGCGGUUUUUAGAUACAUAUGUAUCCGCGAG (((------------------------(((.((((((.(((((((((..((((.(((..(((((...))))).)))..))))..))))))))).)).....)))).))))))........ ( -32.30) >DroPer_CAF1 85540 116 + 1 UGUCGCUGGCUGUGUGCGCGUAUCCGUGUAUCUGUGUGUGGUGCACAUUAUAUUUAGUUACUUGUGUCUGGUUCUACUGCAUUCUAUGCGGCAGUUUCUAGAU----AUGUAUCUGCGAG ..((((((((((.(((((((....))))))).(((((.....))))).......))))))...((((((((..((.((((((...)))))).))...))))))----))......)))). ( -34.60) >consensus UGC________________________AUAGGUGUGAGCGCCGCGCAUUAUAUUUAGUUGCUAGUGCCUGGUUCUACUAUGUUCGGUGCGGCGGCUUUUAGAU____AUGUAUCUGCGAG ......................................((((((((.....((.((((.(((((...)))))...)))).))...)))))))).(((.(((((........))))).))) (-18.57 = -17.68 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:20 2006