
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,598,692 – 9,598,788 |
| Length | 96 |
| Max. P | 0.921606 |

| Location | 9,598,692 – 9,598,788 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
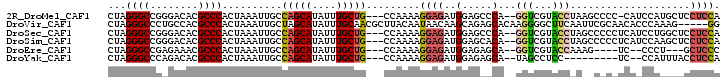
>2R_DroMel_CAF1 9598692 96 + 20766785 CUAGGGCCGGGACACGCCCACUAAAUUGCCAGCAUAUUUGCUG---CCAAAAGGAGAUGGAGCCCA--GGUCGUACCUAAGCCCC-CAUCCAUGCUCCUCCA ...((((.(.....)))))......(((.(((((....)))))---.))).((((((((((((..(--((.....)))..))...-..))))).)))))... ( -31.70) >DroVir_CAF1 14263 97 + 1 CUAGGGCCCUGCCACGCCCACUAAAUUGCUAGCAUAUUUGCAACGCUUACAAUAACAAGCAGAGCACAAGGGGCUUCAAUUCGCAACACCCAAAG-----GG ...(((...(((...((((.((....((((.(((....)))...((((........))))..))))..))))))........)))...)))....-----.. ( -23.00) >DroSec_CAF1 13754 97 + 1 CUAGGGCCGGGACACGCCCACUAAAUUGCCAGCAUAUUUGCUG---CCAAAAGGAGAUGGAGCCCA--GGUCGUACCUAGCCCCCUCAUCCUGGCUCCUCCA ..(((((((((.....)))......(((.(((((....)))))---.))).((((((.((.((..(--((.....))).)).)).)).))))))).)))... ( -34.30) >DroSim_CAF1 22547 97 + 1 CUAGGGCCGGGACACGCCCACUAAAUUGCCAGCAUAUUUGCUG---CCAAAAGGAGAUGGAGCACA--GGUCGUACCUAGCCCCCUCAUCCAAGCUCCUCCA ...((((.(.....)))))......(((.(((((....)))))---.))).(((((.((((....(--((..((.....))..)))..))))..)))))... ( -30.10) >DroEre_CAF1 13968 88 + 1 CUAGGGCCGAGAAACGCCCACUAAAUUGCCAGCAUAUUUGCUG---CCAAAAGGAGAUGGAGAGCA--GGUCGUACCAAAG----UC--CCCU---GCUCCC ...((((........)))).((...(((.(((((....)))))---.)))....))..((.(((((--((.(........)----..--.)))---)))))) ( -28.50) >DroYak_CAF1 23229 86 + 1 CUAGGGCCCAGACACGCCCACUAAAUUGCCAGCAUAUUUGCUG---CCAAAAGGAGAUGGAGAGCA--UAGCCUCC---------UC--CCAUUUACCUCCA ...((((........))))......(((.(((((....)))))---.))).(((((((((.(((..--.......)---------))--)))))).)))... ( -26.20) >consensus CUAGGGCCGGGACACGCCCACUAAAUUGCCAGCAUAUUUGCUG___CCAAAAGGAGAUGGAGAGCA__GGUCGUACCUAACCCCCUCAUCCAUGCUCCUCCA ...((((........))))..........(((((....))))).........((((..(.....)...(((...)))....................)))). (-13.15 = -13.18 + 0.03)

| Location | 9,598,692 – 9,598,788 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.35 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
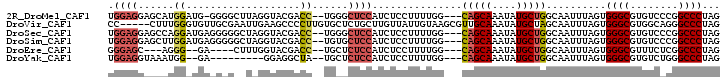
>2R_DroMel_CAF1 9598692 96 - 20766785 UGGAGGAGCAUGGAUG-GGGGCUUAGGUACGACC--UGGGCUCCAUCUCCUUUUGG---CAGCAAAUAUGCUGGCAAUUUAGUGGGCGUGUCCCGGCCCUAG .((.((..((((....-((((((((((.....))--))))))))..(((((.(((.---(((((....))))).)))...)).)))))))..))..)).... ( -40.60) >DroVir_CAF1 14263 97 - 1 CC-----CUUUGGGUGUUGCGAAUUGAAGCCCCUUGUGCUCUGCUUGUUAUUGUAAGCGUUGCAAAUAUGCUAGCAAUUUAGUGGGCGUGGCAGGGCCCUAG ..-----....((((.((((.(......((((((..((((..(((((......)))))...(((....))).))))....)).)))).).)))).))))... ( -31.90) >DroSec_CAF1 13754 97 - 1 UGGAGGAGCCAGGAUGAGGGGGCUAGGUACGACC--UGGGCUCCAUCUCCUUUUGG---CAGCAAAUAUGCUGGCAAUUUAGUGGGCGUGUCCCGGCCCUAG ...(((.(((.((((..((((.(((((.....))--))).))))..(((((.(((.---(((((....))))).)))...)).)))...)))).)))))).. ( -42.90) >DroSim_CAF1 22547 97 - 1 UGGAGGAGCUUGGAUGAGGGGGCUAGGUACGACC--UGUGCUCCAUCUCCUUUUGG---CAGCAAAUAUGCUGGCAAUUUAGUGGGCGUGUCCCGGCCCUAG ...(((.(((.((((...(((((((((.....))--)).)))))..(((((.(((.---(((((....))))).)))...)).)))...)))).)))))).. ( -38.30) >DroEre_CAF1 13968 88 - 1 GGGAGC---AGGG--GA----CUUUGGUACGACC--UGCUCUCCAUCUCCUUUUGG---CAGCAAAUAUGCUGGCAAUUUAGUGGGCGUUUCUCGGCCCUAG ((((((---(((.--.(----(....))....))--))))).))........(((.---(((((....))))).)))......((((........))))... ( -33.80) >DroYak_CAF1 23229 86 - 1 UGGAGGUAAAUGG--GA---------GGAGGCUA--UGCUCUCCAUCUCCUUUUGG---CAGCAAAUAUGCUGGCAAUUUAGUGGGCGUGUCUGGGCCCUAG .((((((....((--((---------(.(.....--).))))).))))))..(((.---(((((....))))).)))......((((.(....).))))... ( -29.60) >consensus UGGAGGAGCAUGGAUGAGGGGCCUAGGUACGACC__UGCGCUCCAUCUCCUUUUGG___CAGCAAAUAUGCUGGCAAUUUAGUGGGCGUGUCCCGGCCCUAG .((((......((...................))......))))...............(((((....)))))..........((((........))))... (-12.82 = -13.29 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:27 2006