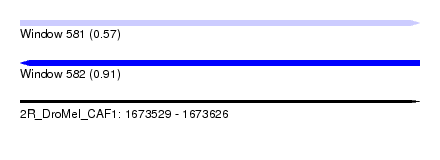
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,673,529 – 1,673,626 |
| Length | 97 |
| Max. P | 0.913048 |

| Location | 1,673,529 – 1,673,626 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1673529 97 + 20766785 AUUCCAUGGGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAACGGUAAUUAUUUU----------------------- ...((....(((((((..((((.........))))((((.(((.....((((.....))))))).))))......)))))))...))..........----------------------- ( -28.90) >DroPse_CAF1 90030 116 + 1 ---GCAACAGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGCCGCGGGUUCGAUCCCCCCAUGGGCCAGGAGUUACUUUUUGGUUUUUUU-UUCAGUGUAAUUUU ---......(((((((.......((.((((...((((..(....)..))))...))))))(((......)))...))))))).((((((((...((((.......-.)))).)))))))) ( -35.70) >DroSec_CAF1 57474 97 + 1 AUUCAAUCGGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAUAGAUUAUUAUUUU----------------------- ....((((.(((((((..((((.........))))((((.(((.....((((.....))))))).))))......)))))))...))))........----------------------- ( -30.90) >DroSim_CAF1 54441 97 + 1 AUUCCAUCGGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCACAGAUUAUUAUUUU----------------------- .....(((.(((((((..((((.........))))((((.(((.....((((.....))))))).))))......)))))))...))).........----------------------- ( -29.40) >DroPer_CAF1 83937 117 + 1 ---GCAACAGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGCCGCGGGUUCGAUCCCCCCAUGGGCCAGGAGUUACUUUUUGGUUUUUUUAUUCAGUGCAAUUUU ---(((...(((((((.......((.((((...((((..(....)..))))...))))))(((......)))...))))))).((((..(((....)))..))))......)))...... ( -34.70) >consensus AUUCCAUCGGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGAGGUUAUUAUUUU_______________________ .........(((((((..((((.........))))((((.(((.....((((.....))))))).))))......)))))))...................................... (-28.04 = -28.04 + -0.00)

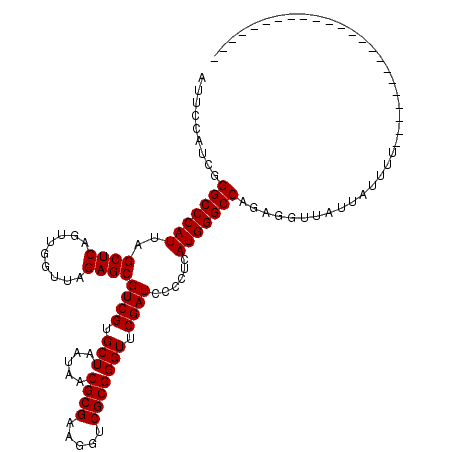
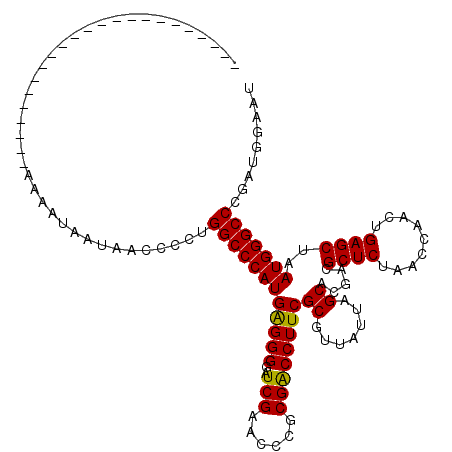
| Location | 1,673,529 – 1,673,626 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -26.88 |
| Energy contribution | -26.40 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1673529 97 - 20766785 -----------------------AAAAUAAUUACCGUUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCGUUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCCCAUGGAAU -----------------------..........((((.(((((((..(((.......)))(((.....)))..............((((.........))))..)))))))..))))... ( -31.00) >DroPse_CAF1 90030 116 - 1 AAAAUUACACUGAA-AAAAAAACCAAAAAGUAACUCCUGGCCCAUGGGGGGAUCGAACCCGCGGCCUUCGCGUUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCUGUUGC--- ..............-..............(((((....((((((((.(((.......))).).....(((.((.....)).))).((((.........))))..))))))).)))))--- ( -31.30) >DroSec_CAF1 57474 97 - 1 -----------------------AAAAUAAUAAUCUAUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCGUUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCCGAUUGAAU -----------------------.......(((((...(((((((..(((.......)))(((.....)))..............((((.........))))..))))))).)))))... ( -29.60) >DroSim_CAF1 54441 97 - 1 -----------------------AAAAUAAUAAUCUGUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCGUUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCCGAUGGAAU -----------------------..........((((((((((((..(((.......)))(((.....)))..............((((.........))))..)))))))..))))).. ( -29.20) >DroPer_CAF1 83937 117 - 1 AAAAUUGCACUGAAUAAAAAAACCAAAAAGUAACUCCUGGCCCAUGGGGGGAUCGAACCCGCGGCCUUCGCGUUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCUGUUGC--- .............................(((((....((((((((.(((.......))).).....(((.((.....)).))).((((.........))))..))))))).)))))--- ( -31.30) >consensus _______________________AAAAUAAUAACCCCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCGUUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCCGAUGGAAU ......................................((((((((((((..(((......))))))))((.......)).....((((.........))))..)))))))......... (-26.88 = -26.40 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:18 2006