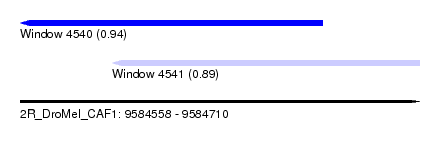
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,584,558 – 9,584,710 |
| Length | 152 |
| Max. P | 0.942994 |

| Location | 9,584,558 – 9,584,673 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -15.83 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
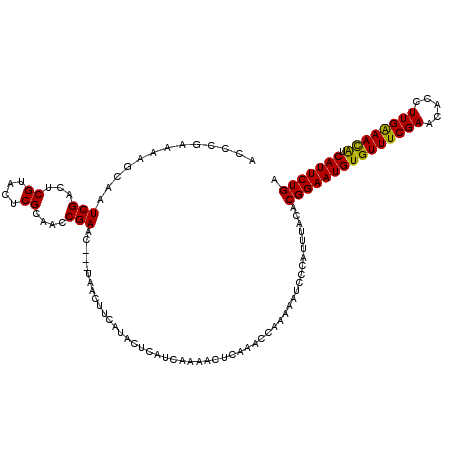
>2R_DroMel_CAF1 9584558 115 - 20766785 AACUUCAUACUCAUCAAAACUCAAAUCAAAAAUCCCAUUUACACGGAAUGUGUUUCGAACACCUUGAAAUAUCAUUCUGAUUUA-----UACUUUCUUAAGUUUCUGUUUCAAUAUUCGU ................((((.......................(((((((((((((((.....)))))))).))))))).....-----.((((....))))....)))).......... ( -14.90) >DroSim_CAF1 8023 115 - 1 AACUUCAUACUCAUUAAAACUCAAACCAAAAAUCCCAUUUACACGGAAUGUGUUUCGAAUACCUUGAAACGUCAUUCUGAUUUA-----UACUUUCUUAAGUUUCUGUUUCAAUAUUCGU ............(((.((((..((((.................(((((((((((((((.....)))))))).)))))))..(((-----........)))))))..)))).)))...... ( -17.90) >DroYak_CAF1 9173 119 - 1 AACUUCAUACUCAUCAAAUCUCAAACCAA-AAUCCCAUUUACACGGAAUGUGUUUCGAACACCUUGGAACACCAUUCUGAUUUCUUAAGUUCUUUCUUAAGUUUCUGUUUCAAUAUUCGU ......................((((...-.............(((((((((((((((.....)))))))).)))))))....((((((......)))))).....)))).......... ( -19.80) >consensus AACUUCAUACUCAUCAAAACUCAAACCAAAAAUCCCAUUUACACGGAAUGUGUUUCGAACACCUUGAAACAUCAUUCUGAUUUA_____UACUUUCUUAAGUUUCUGUUUCAAUAUUCGU ...........................................(((((((((((((((.....)))))))).)))))))......................................... (-15.83 = -15.17 + -0.66)


| Location | 9,584,593 – 9,584,710 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -16.83 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9584593 117 - 20766785 ACCCGAAAAGCAAUCGACUCGUACUCGCAACCGAAC---UAACUUCAUACUCAUCAAAACUCAAAUCAAAAAUCCCAUUUACACGGAAUGUGUUUCGAACACCUUGAAAUAUCAUUCUGA ....(((......(((...((....))....)))..---....))).....................................(((((((((((((((.....)))))))).))))))). ( -15.80) >DroSim_CAF1 8058 120 - 1 AUCCGAAAAGCAAUCGACUCGUACUCGCAACCGAACUACUAACUUCAUACUCAUUAAAACUCAAACCAAAAAUCCCAUUUACACGGAAUGUGUUUCGAAUACCUUGAAACGUCAUUCUGA ...(((.......)))....(((.(((....)))..)))............................................(((((((((((((((.....)))))))).))))))). ( -17.50) >DroYak_CAF1 9213 115 - 1 -CCCGAAAAGCAAUCGACUCGUACUCGCAACCGAAC---UAACUUCAUACUCAUCAAAUCUCAAACCAA-AAUCCCAUUUACACGGAAUGUGUUUCGAACACCUUGGAACACCAUUCUGA -...(((......(((...((....))....)))..---....))).......................-.............(((((((((((((((.....)))))))).))))))). ( -17.20) >consensus ACCCGAAAAGCAAUCGACUCGUACUCGCAACCGAAC___UAACUUCAUACUCAUCAAAACUCAAACCAAAAAUCCCAUUUACACGGAAUGUGUUUCGAACACCUUGAAACAUCAUUCUGA .............(((...((....))....))).................................................(((((((((((((((.....)))))))).))))))). (-16.83 = -16.17 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:17 2006