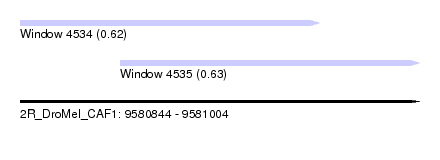
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,580,844 – 9,581,004 |
| Length | 160 |
| Max. P | 0.629342 |

| Location | 9,580,844 – 9,580,964 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -44.37 |
| Consensus MFE | -37.05 |
| Energy contribution | -37.83 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9580844 120 + 20766785 CGCAGCGUGCCACUACCAUGCUUGCUCCUCGCUGUCUUACGUUUCAUCUCAGCCACACUGCCGUACACCAGCGGUCCACUCCUCAUGGGAGAUCCAUUGGAUGCUGGCGGAGUGGCUGCG .((((((((.......))))).)))........................(((((((.((((((((..((((.((....((((.....))))..)).)))).))).))))).))))))).. ( -43.00) >DroSim_CAF1 4251 120 + 1 CGCAGCGUGCCACUACCAUGCUUGCUCCUCGCUGUCUUACGCUUCAUCUCAGCCACACUGCCGUACACCAGCGGUCCACUCCUCAUGGGAGAUCCAUUCUAUGCUGGCGGAGUGGCUGCA .((((((((.......))))).))).....((........)).......(((((((.(((((((......))((....((((.....))))..))..........))))).))))))).. ( -38.30) >DroYak_CAF1 5373 120 + 1 CGAAGCGUGCCACUACCAUGCUUGCUCCUCGCGGUCUUACGCUUCAUUUCAGCCACACUGCCGUACACCAGCGGACCACUCCUCAUGGGAGAUCCAUUGGAUGUAGGCGGAGUGGCUGCA .((((((((((.(......)...((.....)))))...)))))))....(((((((.(((((.((((((((.(((...((((.....)))).))).)))).))))))))).))))))).. ( -51.80) >consensus CGCAGCGUGCCACUACCAUGCUUGCUCCUCGCUGUCUUACGCUUCAUCUCAGCCACACUGCCGUACACCAGCGGUCCACUCCUCAUGGGAGAUCCAUUGGAUGCUGGCGGAGUGGCUGCA .((((((((.......))))).))).....((........)).......(((((((.(((((...((((((.((....((((.....))))..)).)))).))..))))).))))))).. (-37.05 = -37.83 + 0.78)

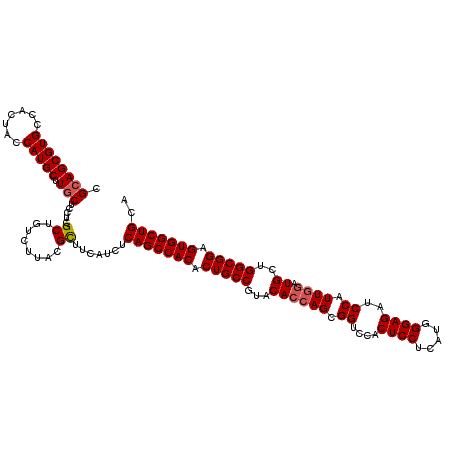
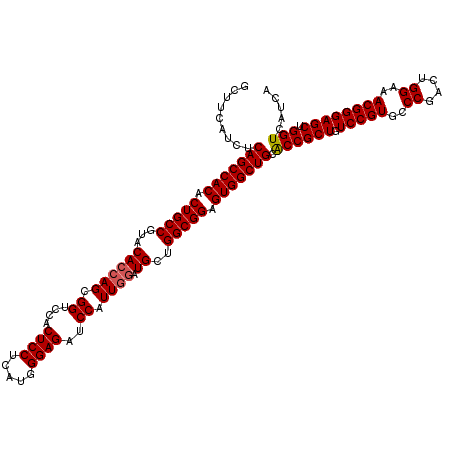
| Location | 9,580,884 – 9,581,004 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -48.77 |
| Consensus MFE | -45.25 |
| Energy contribution | -45.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9580884 120 + 20766785 GUUUCAUCUCAGCCACACUGCCGUACACCAGCGGUCCACUCCUCAUGGGAGAUCCAUUGGAUGCUGGCGGAGUGGCUGCGCCGCUGUCCGUGCCCGACUGGAAACGGGAGCUGGUCAUCA .........(((((((.((((((((..((((.((....((((.....))))..)).)))).))).))))).))))))).((((((.(((((..((....))..)))))))).)))..... ( -49.30) >DroSim_CAF1 4291 120 + 1 GCUUCAUCUCAGCCACACUGCCGUACACCAGCGGUCCACUCCUCAUGGGAGAUCCAUUCUAUGCUGGCGGAGUGGCUGCACCGCUGUCCGUGCCCGACUGGAAACGGGAGCUGGUCAUCA .........(((((((.(((((((......))((....((((.....))))..))..........))))).))))))).((((((.(((((..((....))..)))))))).)))..... ( -43.60) >DroYak_CAF1 5413 120 + 1 GCUUCAUUUCAGCCACACUGCCGUACACCAGCGGACCACUCCUCAUGGGAGAUCCAUUGGAUGUAGGCGGAGUGGCUGCACCGCUGUCCGUGCCCGACUGGAAACGGGAGCUGGUCAUCA .........(((((((.(((((.((((((((.(((...((((.....)))).))).)))).))))))))).))))))).((((((.(((((..((....))..)))))))).)))..... ( -53.40) >consensus GCUUCAUCUCAGCCACACUGCCGUACACCAGCGGUCCACUCCUCAUGGGAGAUCCAUUGGAUGCUGGCGGAGUGGCUGCACCGCUGUCCGUGCCCGACUGGAAACGGGAGCUGGUCAUCA .........(((((((.(((((...((((((.((....((((.....))))..)).)))).))..))))).))))))).((((((.(((((..((....))..)))))))).)))..... (-45.25 = -45.70 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:11 2006