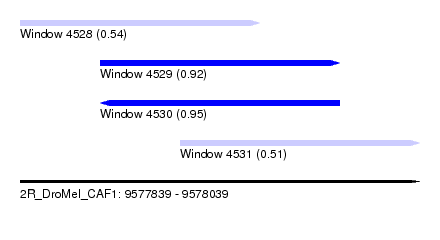
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,577,839 – 9,578,039 |
| Length | 200 |
| Max. P | 0.952124 |

| Location | 9,577,839 – 9,577,959 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -28.61 |
| Energy contribution | -28.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9577839 120 + 20766785 UCCGGCGGCUGCCUUUCAGUUUGUUUUCAAUGGUAGGAUGUUGGGUAUCUGUUAACUAAUCAUUAAAUAUUGCAUACAUACUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGC ..(((((((.((...(((((.(((...(((((....((((...(((........)))...))))...)))))...))).))).((((((((....))))))))))...)).))))).)). ( -32.10) >DroSim_CAF1 1251 116 + 1 UCCGGCGGCUGCCUUUCAGUUUGUUUUCAAUGGUAGGAUGCUGGGUAUCUGUUAACUAAUCAUUAAAUAUUGCAUACA----UGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGC (((((((.(((((..................)))))..))))))).......(((((((((.........((....))----.((((((((....)))))))))))))))))........ ( -31.77) >DroYak_CAF1 2385 120 + 1 AUCGGCUACUGCCUUUCAGUUUGUUUUCAAUGGUAGGAUGCUGGGAAUCUGUUAACUAAUCAUUAAAUAUUGCAUACAUACUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCGCGC .(((((..(((((..................)))))...)))))......(((((((((((.........((....)).....((((((((....)))))))))))))))))..)).... ( -28.87) >consensus UCCGGCGGCUGCCUUUCAGUUUGUUUUCAAUGGUAGGAUGCUGGGUAUCUGUUAACUAAUCAUUAAAUAUUGCAUACAUACUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGC .(((((..(((((..................)))))...)))))........(((((((((.........((....)).....((((((((....)))))))))))))))))........ (-28.61 = -28.17 + -0.44)

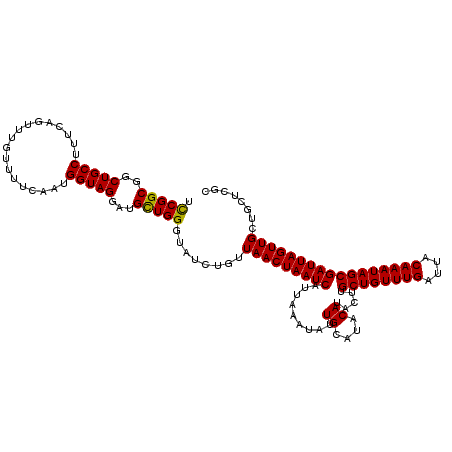
| Location | 9,577,879 – 9,577,999 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9577879 120 + 20766785 UUGGGUAUCUGUUAACUAAUCAUUAAAUAUUGCAUACAUACUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUA .(((((((..(((((((((((.........((....)).....((((((((....))))))))))))))))((((((....)))))).......)))..))))).))............. ( -29.70) >DroSim_CAF1 1291 116 + 1 CUGGGUAUCUGUUAACUAAUCAUUAAAUAUUGCAUACA----UGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUA ...(((((..(((((((((((.........((....))----.((((((((....))))))))))))))))((((((....)))))).......)))..)))))................ ( -29.60) >DroYak_CAF1 2425 120 + 1 CUGGGAAUCUGUUAACUAAUCAUUAAAUAUUGCAUACAUACUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCGCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUA ...........................((((((........((((((((((....))))))))))...((.(((((......)))))..)).................))))))...... ( -26.60) >consensus CUGGGUAUCUGUUAACUAAUCAUUAAAUAUUGCAUACAUACUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUA .(((((((..(((((((((((.........((....)).....((((((((....))))))))))))))))((((((....)))))).......)))..))))).))............. (-26.60 = -27.27 + 0.67)

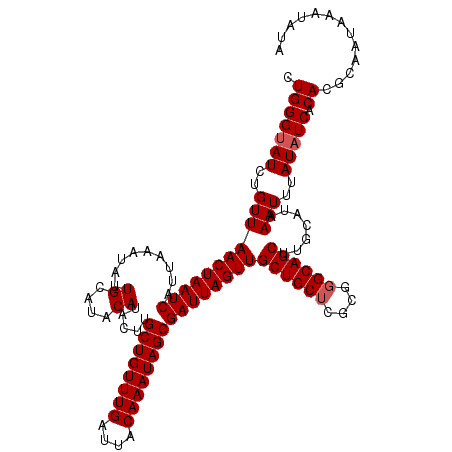

| Location | 9,577,879 – 9,577,999 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -27.07 |
| Energy contribution | -27.07 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9577879 120 - 20766785 UAUAUUUAUUGCGUGUGAUAUAAAUUUAUGCAAGCUGCCCGCGAGCAGCAACUAAUCGCUAUUUGUAAUCAAACAGCAAGUAUGUAUGCAAUAUUUAAUGAUUAGUUAACAGAUACCCAA ..(((((.(((((((...........)))))))(((((......)))))(((((((((((.((((....)))).)))((((((.......))))))...))))))))...)))))..... ( -29.30) >DroSim_CAF1 1291 116 - 1 UAUAUUUAUUGCGUGUGAUAUAAAUUUAUGCAAGCUGCCCGCGAGCAGCAACUAAUCGCUAUUUGUAAUCAAACAGCA----UGUAUGCAAUAUUUAAUGAUUAGUUAACAGAUACCCAG ..(((((.(((((((...........)))))))(((((......)))))(((((((((((.((((....)))).))).----((....)).........))))))))...)))))..... ( -27.30) >DroYak_CAF1 2425 120 - 1 UAUAUUUAUUGCGUGUGAUAUAAAUUUAUGCAAGCUGCCCGCGCGCAGCAACUAAUCGCUAUUUGUAAUCAAACAGCAAGUAUGUAUGCAAUAUUUAAUGAUUAGUUAACAGAUUCCCAG ........(((((((...........)))))))(((((......)))))(((((((((((.((((....)))).)))((((((.......))))))...))))))))............. ( -29.10) >consensus UAUAUUUAUUGCGUGUGAUAUAAAUUUAUGCAAGCUGCCCGCGAGCAGCAACUAAUCGCUAUUUGUAAUCAAACAGCAAGUAUGUAUGCAAUAUUUAAUGAUUAGUUAACAGAUACCCAG ........(((((((...........)))))))(((((......)))))(((((((((((.((((....)))).))).....((....)).........))))))))............. (-27.07 = -27.07 + -0.00)

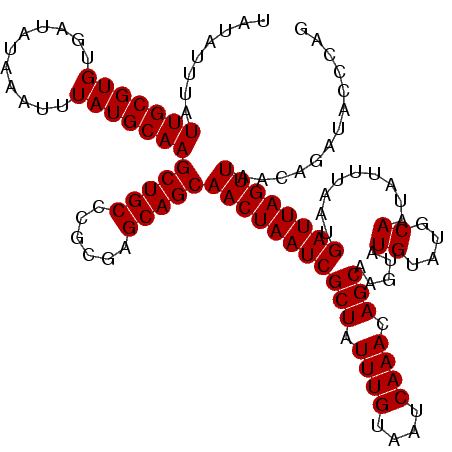

| Location | 9,577,919 – 9,578,039 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -24.79 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9577919 120 + 20766785 CUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUAAUAAAAGUAACUUAAGAUUUCGAAAUCAAGAAAAUUCUAA .((((((((((....))))))))))..((((((((((....))))))((((.................))))................))))..(((((((........))))..))).. ( -24.43) >DroSim_CAF1 1329 118 + 1 --UGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUAAUAAAAGUAACAUAAGAUUUCGAAAUCAAGAAAAUUCUAA --.((((((((....)))))))).....(((((((((....))))))((((.................))))................)))...(((((((........))))..))).. ( -23.23) >DroYak_CAF1 2465 120 + 1 CUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCGCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUAAUAAAAGUAACUUAGGAUUUCGAAAUCAAGAAAAUUCUAA .((((((((((....))))))))))..((((((((((.((.....)).)))....((((((..........))))))........)))))))(((((((((........).)))))))). ( -26.70) >consensus CUUGCUGUUUGAUUACAAAUAGCGAUUAGUUGCUGCUCGCGGGCAGCUUGCAUAAAUUUAUAUCACACGCAAUAAAUAUAAUAAAAGUAACUUAAGAUUUCGAAAUCAAGAAAAUUCUAA .((((((((((....))))))))))...((((((((..((.....))..))....((((((..........))))))........))))))...(((((((........))))..))).. (-21.79 = -21.90 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:07 2006