
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,558,687 – 9,558,814 |
| Length | 127 |
| Max. P | 0.897724 |

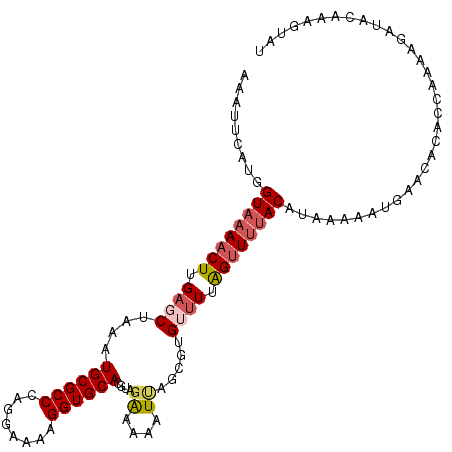
| Location | 9,558,687 – 9,558,784 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9558687 97 - 20766785 AAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUACAAAAAUUAGCUUGUUUUAGUUUUACAU------------GCCAAAAGAUGCAAAGUAU .........((((((((.(((((((.((((((........))))))..........))))))).....)))))))).(------------((........)))...... ( -21.00) >DroSec_CAF1 35255 109 - 1 AAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUAGAGAAAUUAGAGUGUUUUAGUUUUACAUAAAAAUUUACACACCAAAAGAUUCAAAGUAU .......((((..........(((..((((((........))))))..)))..........((((...((((((.....)))))).))))))))............... ( -20.90) >DroSim_CAF1 35098 109 - 1 AAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUAGAAAAAUUAGAGUGUUUUAGUUUUACAUACAAAUUUACACACCAAAAGGUACAAAGUAU .........((((((((....(((..((((((........))))))..))).(((((......)))))))))))))..............(((....)))......... ( -20.80) >DroEre_CAF1 37491 106 - 1 AAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAGGGUGCACGGUGAAAAAUCAACAUGGAUUAGUUGCACAUAAAGGUGAACAUACCAAA--AAAC-AUAUAC .((((((((...........(((...(((((((......))))))).)))..........)))))))).(((.(((......))))))........--....-...... ( -24.80) >DroYak_CAF1 36673 108 - 1 AAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGGUGGAAAAUCAACUUGGUUUGGAUUUACAUAAAAAUGAACAUAGCAUAGAAAAC-AAGUAU ...(((((.(((((..(..((((((.((((((........)))))).(((.....)))...))))))..).)))))......)))))...............-...... ( -20.10) >consensus AAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUAGAAAAAUUAGCGUGUUUUAGUUUUACAUAAAAAUGAACACACCAAAAGAUACAAAGUAU .........((((((((.((((....((((((........))))))....((....)).....)))).))))))))................................. (-13.86 = -14.78 + 0.92)

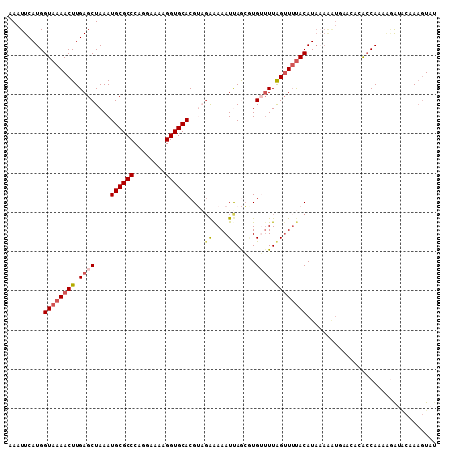
| Location | 9,558,715 – 9,558,814 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9558715 99 + 20766785 UAAAACAAGCUAAUUUUUGUACGUGCACCUUUUCCUGGGCGCAUUUAGCUCAAGUUUUACCAUGAAUUUCGUUCAUGCAUUAACCCUCGGUUGCCUGAG ((((((.((((((.........((((.(((......))).))))))))))...)))))).(((((((...)))))))........(((((....))))) ( -23.10) >DroSec_CAF1 35295 99 + 1 UAAAACACUCUAAUUUCUCUACGUGCACCUUUUCCUGGGCGCAUUUAGCUCAAGUUUUACCAUGAAUUUCGUUCAUGCAUUAACCCUCGGUUGCCUGAG ..................(((.((((.(((......))).)))).))).....(((....(((((((...)))))))....))).(((((....))))) ( -19.60) >DroSim_CAF1 35138 99 + 1 UAAAACACUCUAAUUUUUCUACGUGCACCUUUUCCUGGGCGCAUUUAGCUCAAGUUUUACCAUGAAUUUCGUUCAUGCAUUAACCCUCGGUUGCCUGAG ..................(((.((((.(((......))).)))).))).....(((....(((((((...)))))))....))).(((((....))))) ( -19.60) >DroEre_CAF1 37528 99 + 1 UAAUCCAUGUUGAUUUUUCACCGUGCACCCUUUCCUGGGCGCAUUUAGCUCAAGUUUUACCAUGAAUUUCGUUCAUGCAUUAACCCUCGGUUGCCUGAG ((((.((((.(((...((((..((((.(((......))).))))..(((....)))......))))..)))..)))).))))...(((((....))))) ( -21.50) >DroYak_CAF1 36712 99 + 1 CAAACCAAGUUGAUUUUCCACCGUGCACCUUUUCCUGGGCGCAUUUAGCUCAAGUUUUACCAUGAAUUUCGUUCAUGCAUUAACCCUCGGUUGCCUGAG ..((((.((((((.........((((.(((......))).))))))))))...(((....(((((((...)))))))....)))....))))....... ( -19.60) >consensus UAAAACAAGCUAAUUUUUCUACGUGCACCUUUUCCUGGGCGCAUUUAGCUCAAGUUUUACCAUGAAUUUCGUUCAUGCAUUAACCCUCGGUUGCCUGAG ........(((((.........((((.(((......))).)))))))))....(((....(((((((...)))))))....))).(((((....))))) (-18.34 = -18.10 + -0.24)


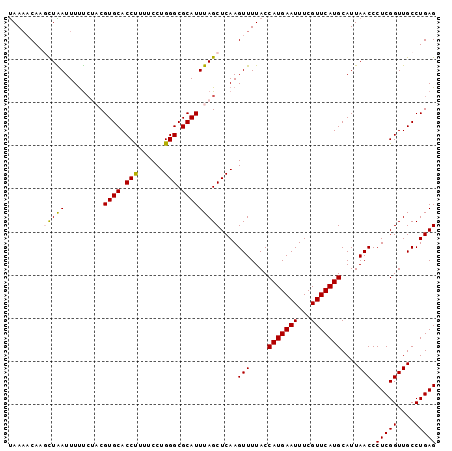
| Location | 9,558,715 – 9,558,814 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9558715 99 - 20766785 CUCAGGCAACCGAGGGUUAAUGCAUGAACGAAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUACAAAAAUUAGCUUGUUUUA ..(((((...((((..(((...((((((.....)))))).)))..)))).......((((((........)))))).............)))))..... ( -24.90) >DroSec_CAF1 35295 99 - 1 CUCAGGCAACCGAGGGUUAAUGCAUGAACGAAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUAGAGAAAUUAGAGUGUUUUA (((.......((((..(((...((((((.....)))))).)))..))))..(((..((((((........))))))..)))........)))....... ( -24.20) >DroSim_CAF1 35138 99 - 1 CUCAGGCAACCGAGGGUUAAUGCAUGAACGAAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUAGAAAAAUUAGAGUGUUUUA (((.......((((..(((...((((((.....)))))).)))..))))..(((..((((((........))))))..)))........)))....... ( -24.20) >DroEre_CAF1 37528 99 - 1 CUCAGGCAACCGAGGGUUAAUGCAUGAACGAAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAGGGUGCACGGUGAAAAAUCAACAUGGAUUA .((((((...((((..(((...((((((.....)))))).)))..)))).)))...(((((((......)))))))...)))..((((......)))). ( -26.30) >DroYak_CAF1 36712 99 - 1 CUCAGGCAACCGAGGGUUAAUGCAUGAACGAAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGGUGGAAAAUCAACUUGGUUUG .......(((((((((((....((((((.....))))))....))))(((.(((..((((((........))))))...)))....))).))))))).. ( -25.40) >consensus CUCAGGCAACCGAGGGUUAAUGCAUGAACGAAAUUCAUGGUAAAACUUGAGCUAAAUGCGCCCAGGAAAAGGUGCACGUAGAAAAAUUAGCGUGUUUUA ....(((...((((..(((...((((((.....)))))).)))..)))).)))...((((((........))))))....................... (-20.98 = -20.98 + 0.00)

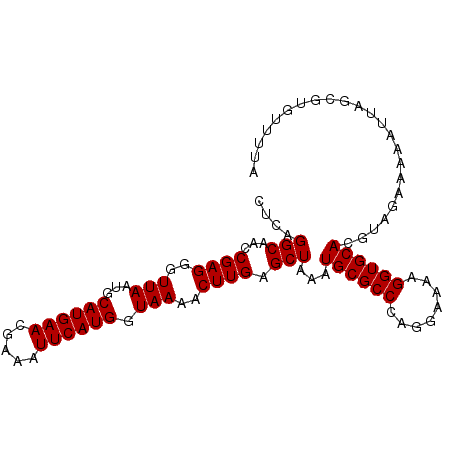
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:01 2006