
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,552,863 – 9,552,953 |
| Length | 90 |
| Max. P | 0.614031 |

| Location | 9,552,863 – 9,552,953 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
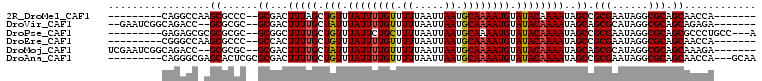
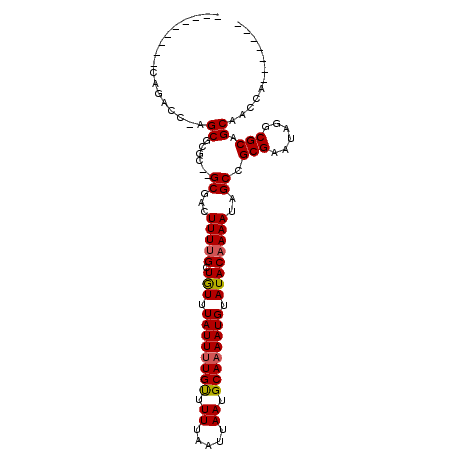
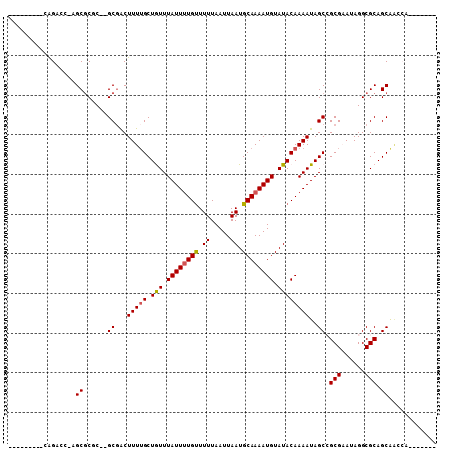
>2R_DroMel_CAF1 9552863 90 - 20766785 ---------CAGGCCAAGCGCCC--GCGACUUUAGCUGUUUAUUUUGUUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCCGCGAAUAGGCGCAGCAACCA------- ---------...((...((((((--(((.((.....(((.((((((((.((.....)).))))))))...)))....)).))))....))))).)).....------- ( -23.00) >DroVir_CAF1 151979 95 - 1 --GAAUCGGCAGACC--GCGCGC--GCGACUUUUGCUAUUUAUUUUGUUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCAGCGCAUAGGCGCAGCAGAGA------- --...((((....))--((((((--((..((((((.(((.((((((((.((.....)).)))))))).)))))))..))..))))....))))....))..------- ( -23.40) >DroPse_CAF1 48360 94 - 1 ---------GAGAGCGCGCGCGC--GCGGCUUUUGCUGUUUAUUCUGCUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCCGCGAAUAGGCGCAGCGCCCUGCC---A ---------.((.((((((((.(--((((((((((.(((.((((.(((...........))).)))).)))))))..))))))).....)))).)))).))...---. ( -34.90) >DroEre_CAF1 31594 90 - 1 ---------CGGGCCAAGCGCCC--GCCACUUUUGCUGUUUAUUUUGUUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCCGCGAAUAGGCGCAGCAACCA------- ---------(((((.....))))--)......(((((((.((((((((.........((((.....))))))))))))(((.......))))))))))...------- ( -26.50) >DroMoj_CAF1 160249 97 - 1 UCGAAUCGGCAGACC--GCGCGC--GCGACUUUUGCUAUUUAUUUUGUUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCAGCGCAUAGGCGCAGCAAAGA------- (((...((((.....--)).)).--.))).(((((((..(((((((((.........((((.....)))))))))))))..((((....))))))))))).------- ( -23.20) >DroAna_CAF1 24876 96 - 1 ---------CAGGGCGAGCACUCGCGCGACUUUUGCUGUUUAUUUUGUUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCCGCGAAUAGGCGCAGCAACCA---GCAA ---------..((((((....)))).......(((((((.((((((((.........((((.....))))))))))))(((.......)))))))))))).---.... ( -26.30) >consensus _________CAGACC_AGCGCGC__GCGACUUUUGCUGUUUAUUUUGUUUUUAAUUAAUGCAAAAUGUAUACAAAAUAGCCGCGAAUAGGCGCAGCAACCA_______ .................((......((...(((((.(((.((((((((.((.....)).)))))))).))))))))..)).(((......))).))............ (-16.23 = -16.20 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:54 2006