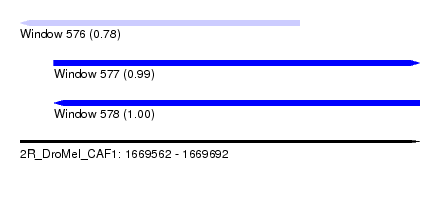
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,669,562 – 1,669,692 |
| Length | 130 |
| Max. P | 0.995800 |

| Location | 1,669,562 – 1,669,653 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1669562 91 - 20766785 CCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCAGUUAAGCACACAAAAAAAUUA ((((((((.......))))).....((((((.....)))))).((((.........))))..)))..((......)).............. ( -25.20) >DroPse_CAF1 84213 90 - 1 CCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCUCUGAAA-CAUCGUAAAAUUC ((((((((.......))))).....((((((.....)))))).((((.........))))..)))............-............. ( -24.00) >DroPer_CAF1 78143 90 - 1 CCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCUCUGAAA-CAUCGUAAAAUUC ((((((((.......))))).....((((((.....)))))).((((.........))))..)))............-............. ( -24.00) >consensus CCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCUCUGAAA_CAUCGUAAAAUUC ((((((((.......))))).....((((((.....)))))).((((.........))))..))).......................... (-24.00 = -24.00 + 0.00)

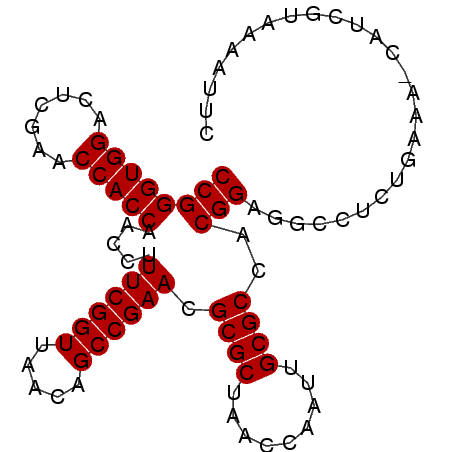
| Location | 1,669,573 – 1,669,692 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.67 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1669573 119 + 20766785 UGUGCUUAACUGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGCAUACGAAAU-CAAAAUUUUUGCUAUGCGUUCUCC .(((((.(((((((.....)))......))))))))).((((((..((((((....))))))..).))))).......((((((((((((((((.-......))))).))))))))))). ( -46.10) >DroPse_CAF1 84224 108 + 1 UG-UUUCAGAGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGU--UAUAA-------CUUUUUUGUAUUCUCUUU-- ..-....(((((((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))..--(((((-------.....)))))..))))..-- ( -39.70) >DroEre_CAF1 60507 107 + 1 CUUAUUUAAGGGCCUCCGUGGCGCAAUUGAUUAGCGUGUUCGACUGUUAACCAACAUUUUGGUGGUUCCAGUCCACCCGGGGGCGCGUAUCAAUUUCGGGGUUUUUU------------- .......(((((((.(((......(((((((..((((.((((..((((....))))....(((((.......))))))))).))))..))))))).)))))))))).------------- ( -32.40) >DroPer_CAF1 78154 108 + 1 UG-UUUCAGAGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGU--AACAA-------CUUUUUUGUAUUCUCUUU-- ..-....(((((((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))..--.((((-------.....))))...))))..-- ( -40.50) >consensus UG_UUUCAAAGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGC__AACAA_______CUUUUUUGUAUUCUCUUU__ ...........(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).................................... (-32.30 = -32.67 + 0.38)

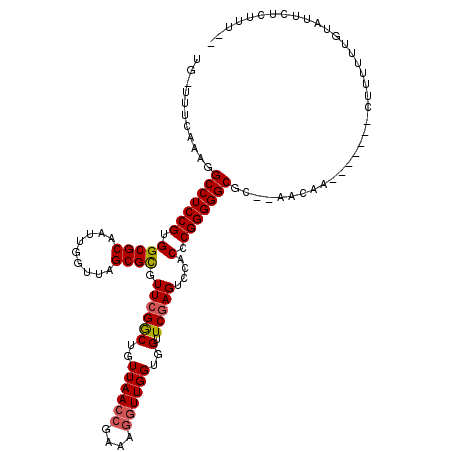
| Location | 1,669,573 – 1,669,692 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -26.98 |
| Energy contribution | -26.85 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
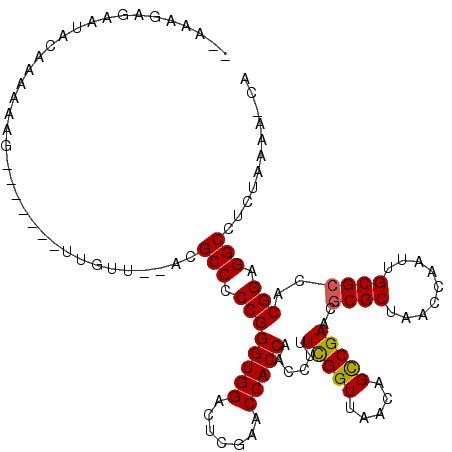
>2R_DroMel_CAF1 1669573 119 - 20766785 GGAGAACGCAUAGCAAAAAUUUUG-AUUUCGUAUGCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCAGUUAAGCACA .(..((((((((..(((.......-.)))..)))))(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).)))...)... ( -36.30) >DroPse_CAF1 84224 108 - 1 --AAAGAGAAUACAAAAAAG-------UUAUA--ACGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCUCUGAAA-CA --..((((...........(-------(....--))(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))))))....-.. ( -33.10) >DroEre_CAF1 60507 107 - 1 -------------AAAAAACCCCGAAAUUGAUACGCGCCCCCGGGUGGACUGGAACCACCAAAAUGUUGGUUAACAGUCGAACACGCUAAUCAAUUGCGCCACGGAGGCCCUUAAAUAAG -------------......(((((.(((((((...((((....))))(((((.(((((.(.....).)))))..)))))..........)))))))......))).))............ ( -27.10) >DroPer_CAF1 78154 108 - 1 --AAAGAGAAUACAAAAAAG-------UUGUU--ACGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCUCUGAAA-CA --..((((...((((.....-------)))).--..(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))))))....-.. ( -34.00) >consensus __AAAGAGAAUACAAAAAAG_______UUGUU__ACGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCUCUAAAA_CA ....................................(((.((((((((.......)))))......(((((.....)))))..((((.........))))..))).)))........... (-26.98 = -26.85 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:14 2006