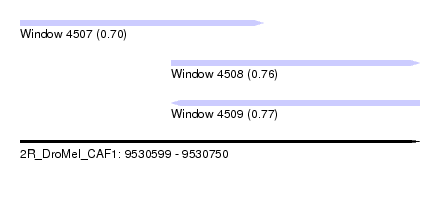
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,530,599 – 9,530,750 |
| Length | 151 |
| Max. P | 0.769300 |

| Location | 9,530,599 – 9,530,691 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9530599 92 + 20766785 ------AUAGCAAAUGAUGUGCGGGAUAUAUAGA--------CGCCGGCAUAAAUUAUUGCCGAGCUUAACGGGAAUAAUAUUGUUGCAAAAGGCAAUUC-----GUACGA ------...........(((((((..........--------.(((((((........))))).)).................(((((.....)))))))-----))))). ( -22.00) >DroSec_CAF1 7600 100 + 1 ------AUAGCAAAUGAUGUGCGGGAUAUAUAGAUAUAUAGCUGCCGGCAUAAAUUAUUGCCGGGCUUAACGGGAAUAAUAUUGUUGCAAAAGGCAAUUC-----GUACGA ------...........((((((((.(((((....)))))..((((.((((((((((((.(((.......))).)))))).))).)))....)))).)))-----))))). ( -27.10) >DroSim_CAF1 6254 100 + 1 ------AUAGCAAAUGAUGUGCGGGAUAUAUAGAUAUAUAGCCGACGGCAUAAAUUAUUGCCGGGCUUAACGGGAAUAAUAUUGUUGCAAAAGGCAAUUC-----GUACGA ------...........((((((((.(((((....)))))(((....((((((((((((.(((.......))).)))))).))).)))....)))..)))-----))))). ( -27.50) >DroEre_CAF1 9151 102 + 1 CGGUAUAUAACAAAUGAUGUGCGGGAUA--------UAUAGCAGCCGGCAUAAAUUAUUGCCGCGCUUU-CUGGCAUAAUAUUGUUGCAAAAGGCAAUGCAAUUUCUUCGA (((.........((((.(((((.(((..--------...(((.((.((((........))))))))).)-)).))))).))))((((((........))))))....))). ( -26.70) >consensus ______AUAGCAAAUGAUGUGCGGGAUAUAUAGA__UAUAGCCGCCGGCAUAAAUUAUUGCCGGGCUUAACGGGAAUAAUAUUGUUGCAAAAGGCAAUUC_____GUACGA .......((((((....(((((.((...................)).))))).((((((.(((.......))).)))))).))))))........................ (-15.07 = -15.63 + 0.56)

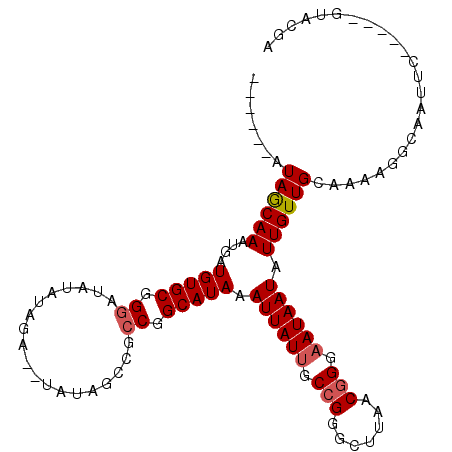
| Location | 9,530,656 – 9,530,750 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -15.24 |
| Energy contribution | -16.24 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9530656 94 + 20766785 GGGAAUAAUAUUGUUGCAAAAGGCAAUUC-----GUACGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGCCAAUAAGCUGAAGCUGGGCCAAGAAUU (((((((((...(((((.....)))))..-----.....))))))))).................((((((((....(((....))).))))))))... ( -27.20) >DroSec_CAF1 7665 91 + 1 GGGAAUAAUAUUGUUGCAAAAGGCAAUUC-----GUACGAUUA---CUUAUCAUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUU .(.(((((....(((((.....)))))..-----....(((..---...)))..))))).)...(((((((((...((((....))))))))))))).. ( -24.00) >DroSim_CAF1 6319 94 + 1 GGGAAUAAUAUUGUUGCAAAAGGCAAUUC-----GUACGAUUAUUUCUUAUCGUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGUUUGGCCAAGAUUU .(.((((....)))).).....(((((..-----..(((((........)))))..)))))...(((((((((((...........))))))))))).. ( -26.20) >DroEre_CAF1 9213 98 + 1 UGGCAUAAUAUUGUUGCAAAAGGCAAUGCAAUUUCUUCGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGU-AAUGAAUCGGGGCUCAGCCAAGACUU ((((........((((((........))))))..((((((((.....((((((...............))))-))..))))))))....))))...... ( -19.56) >DroYak_CAF1 8336 94 + 1 UGGCAUAAUAU-GUUGCAAAAGGCAAUGCAAAUUCUUCAAU---UCCUUAUCAUUUAUUGCAUCGUCUUGGU-AAUGAAUUGCAGCUCAGCCAAGAGUU ((((.......-(((((.....))))).......((.((((---((.((((((...............))))-)).)))))).))....))))...... ( -19.56) >consensus GGGAAUAAUAUUGUUGCAAAAGGCAAUUC_____GUACGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUU .(.(((((....(((((.....)))))...........(((........)))..))))).)....((((((((...((((....))))))))))))... (-15.24 = -16.24 + 1.00)


| Location | 9,530,656 – 9,530,750 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -12.77 |
| Energy contribution | -14.05 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9530656 94 - 20766785 AAUUCUUGGCCCAGCUUCAGCUUAUUGGCCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGUAC-----GAAUUGCCUUUUGCAACAAUAUUAUUCCC ...((((((((.(((....)))....))))))))....(((((..(((((.(....).)))))..-----..)))))...................... ( -21.40) >DroSec_CAF1 7665 91 - 1 AAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAUGCAAUAAAUGAUAAG---UAAUCGUAC-----GAAUUGCCUUUUGCAACAAUAUUAUUCCC ..(((((((((((...........)))))))))))..(((((.....(((..(---((....)))-----..))).....))))).............. ( -21.80) >DroSim_CAF1 6319 94 - 1 AAAUCUUGGCCAAACUUCAACUCAUUGGCCAAGAUGAUGCAAUAAACGAUAAGAAAUAAUCGUAC-----GAAUUGCCUUUUGCAACAAUAUUAUUCCC ..(((((((((((...........)))))))))))...(((((..(((((.(....).)))))..-----..)))))...................... ( -23.60) >DroEre_CAF1 9213 98 - 1 AAGUCUUGGCUGAGCCCCGAUUCAUU-ACCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGAAGAAAUUGCAUUGCCUUUUGCAACAAUAUUAUGCCA ..(((((((.((((......))))..-.)))))))((((((((...((((.(....).)))).....))))))))((.....))............... ( -17.80) >DroYak_CAF1 8336 94 - 1 AACUCUUGGCUGAGCUGCAAUUCAUU-ACCAAGACGAUGCAAUAAAUGAUAAGGA---AUUGAAGAAUUUGCAUUGCCUUUUGCAAC-AUAUUAUGCCA ......(((((((..(((((......-...(((.((((((((.............---..........)))))))).))))))))..-...))).)))) ( -16.40) >consensus AAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGUAC_____GAAUUGCCUUUUGCAACAAUAUUAUUCCC ..(((((((((((...........)))))))))))..(((((...(((((........))))).................))))).............. (-12.77 = -14.05 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:46 2006