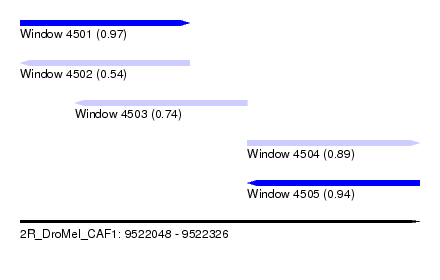
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,522,048 – 9,522,326 |
| Length | 278 |
| Max. P | 0.970155 |

| Location | 9,522,048 – 9,522,166 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -42.95 |
| Consensus MFE | -39.01 |
| Energy contribution | -39.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9522048 118 + 20766785 GGUUGUUGGUUGUUAGUUACUUGGCUAGUUA--GUUGGCUUGUCGCUUUCGUCACUGGAGGCGGCACCAUUUUGUUUGGCGUCGUUUUCUCAUGGGCCAACAGACGUUUCAGCCAGCCGA ....((((((((.(((((....)))))(((.--(((((((..(.(((((((....)))))))(((.(((.......))).)))........)..))))))).)))....))))))))... ( -42.50) >DroSec_CAF1 18647 120 + 1 CGUUGUUGCUUGUUAGUAACUUGGCUAGUUAGAGUUGGCUUGUCGCUUUCGUCAACGGAGGCGGCACCAUUUUGUUUGGCGUCGUUUUCUCAUGGGCCAACAGACGUUUCAGCCAGCCGA ....((((((....))))))((((((.(((.(((.(((..(((((((((((....)))))))))))))).((((((.(((.((((......)))))))))))))...)))))).)))))) ( -43.30) >DroSim_CAF1 19012 120 + 1 CGUUGUUGGCUGUUAGUAACUUGGCUAGUUAGAGUUGGCUUGUCGCUUUCGUCACCGGAGGCGGCACCAUUUUGUUUGGCGUCGUUUUCUCAUGGGCCAACAGACGUUUCAGCCAGCCGA ....((((((((...((...((((((.((.((((.((((.(((((((((((....)))))))))))(((.......))).))))..)))).)).))))))...))....))))))))... ( -47.30) >DroYak_CAF1 18389 112 + 1 ------UGGUUGUUAGUAACUUGGCUAGUUA--GUUGGCUUGUCGCUUUCGUCACUGGAGGCGGCACCAUUUUGUUUGGCGUCGUUUUCUCAUGGGCCAACAGACGUUUCAGCCAGCCGA ------((((((.((((......))))(((.--(((((((..(.(((((((....)))))))(((.(((.......))).)))........)..))))))).)))....))))))..... ( -38.70) >consensus CGUUGUUGGUUGUUAGUAACUUGGCUAGUUA__GUUGGCUUGUCGCUUUCGUCACCGGAGGCGGCACCAUUUUGUUUGGCGUCGUUUUCUCAUGGGCCAACAGACGUUUCAGCCAGCCGA ....(((((((((((((......))))).....(((((((..(.(((((((....)))))))(((.(((.......))).)))........)..)))))))........))))))))... (-39.01 = -39.33 + 0.31)

| Location | 9,522,048 – 9,522,166 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9522048 118 - 20766785 UCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCAGUGACGAAAGCGACAAGCCAAC--UAACUAGCCAAGUAACUAACAACCAACAACC ...((((((((....((..((((((....(((....((.(((((.......))))).)).)))..((..(....)..))..))))))--..))))))).))).................. ( -29.20) >DroSec_CAF1 18647 120 - 1 UCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCGUUGACGAAAGCGACAAGCCAACUCUAACUAGCCAAGUUACUAACAAGCAACAACG ..((((((..((....)).((((((....(((....((.(((((.......))))).)).))).((((.(....)))))..)))))).....))))))..(((.((....)).))).... ( -32.70) >DroSim_CAF1 19012 120 - 1 UCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCGGUGACGAAAGCGACAAGCCAACUCUAACUAGCCAAGUUACUAACAGCCAACAACG ...(.((((((........((((((..............(((((.......))))).(((...((....))...)))....))))))..(((((.....)))))....)))))).).... ( -33.60) >DroYak_CAF1 18389 112 - 1 UCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCAGUGACGAAAGCGACAAGCCAAC--UAACUAGCCAAGUUACUAACAACCA------ ..(((((((((....((..((((((....(((....((.(((((.......))))).)).)))..((..(....)..))..))))))--..))))))).))))...........------ ( -29.60) >consensus UCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCAGUGACGAAAGCGACAAGCCAAC__UAACUAGCCAAGUUACUAACAACCAACAACG ..((((((..((....)).((((((....(((....((.(((((.......))))).)).)))...((.(....)))....)))))).....))))))...................... (-26.20 = -26.20 + -0.00)

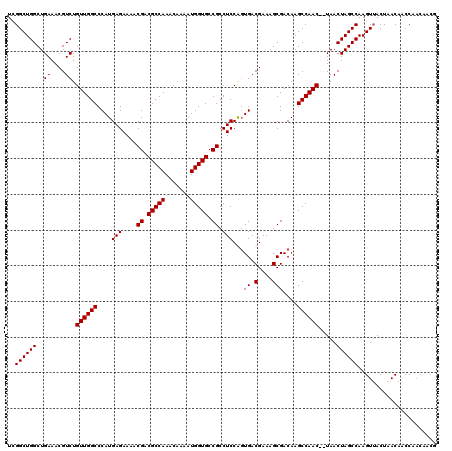
| Location | 9,522,086 – 9,522,206 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -37.67 |
| Energy contribution | -37.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9522086 120 - 20766785 UUAUUGCAAAUGGAGAAGCGGCGAAGAAGGCUGAAAGUCGUCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCAGUGACGAAAGCGACA ..........(((((..(((((..............(((((.(((..((.((....)).))..))).........)))))((((.......))))))))))))))((..(....)..)). ( -36.90) >DroSec_CAF1 18687 120 - 1 UUAUUGCAAACGGAGAAGCGGCGAAGAAGGCUGAAAGUCGUCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCGUUGACGAAAGCGACA ...........((((..(((((..............(((((.(((..((.((....)).))..))).........)))))((((.......)))))))))))))((((.(....))))). ( -39.80) >DroSim_CAF1 19052 120 - 1 UUAUUGCAAACGGAGAAGCGGCGAAGAUGGCUGAAAGUCGUCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCGGUGACGAAAGCGACA ...((((....((((..(((((...((((((.....))))))(((..((.((....)).))..)))..............((((.......)))))))))))))(....)....)))).. ( -42.20) >DroYak_CAF1 18421 120 - 1 UUAUUGCAAAUGGAGAAGCGGCGAAGAUGGCUGAAAGUCGUCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCAGUGACGAAAGCGACA ..........(((((..(((((...((((((.....))))))(((..((.((....)).))..)))..............((((.......))))))))))))))((..(....)..)). ( -39.90) >consensus UUAUUGCAAACGGAGAAGCGGCGAAGAAGGCUGAAAGUCGUCGGCUGGCUGAAACGUCUGUUGGCCCAUGAGAAAACGACGCCAAACAAAAUGGUGCCGCCUCCAGUGACGAAAGCGACA ..........(((((..(((((...((((((.....))))))(((..((.((....)).))..)))..............((((.......)))))))))))))).((.(....)))... (-37.67 = -37.67 + 0.00)

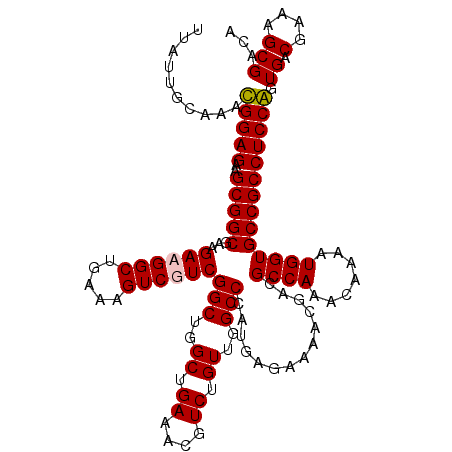
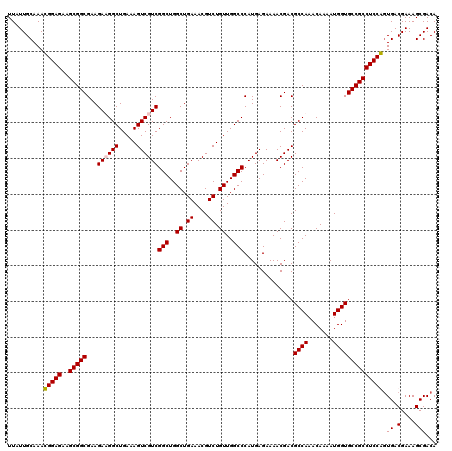
| Location | 9,522,206 – 9,522,326 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -31.94 |
| Energy contribution | -32.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9522206 120 + 20766785 AAUGUAUCUUCUCGCCGGUCGCGUUUAAAACCGCAUUUUUUUCGCCUUGCGUUUGGAAACGCGAGCGGUUUUUGCAGCUAUCUAAUAAAUUCAUUAAUUCCAAUGCUGACGGGCGUUUUA ............((((.((((((((.......(((......((((.(((((((....)))))))))))....))).......((((......)))).....))))).))).))))..... ( -31.30) >DroSec_CAF1 18807 120 + 1 AAUGUAUGUUCUCGCCGGUCGCGUUUAAAACCGCAUUUUUUUCGCCUUGCGUUUGGAAACGCGAGCGGUUUUUGCAGCUACCAAAUUAAUUCAUUAAUUCCCAUGCUGACGGGCGUUUUA .....((((((..((((.((((((((..((.((((............)))).))..)))))))).)))).....((((.....((((((....)))))).....))))..)))))).... ( -35.30) >DroSim_CAF1 19172 120 + 1 AAUGUAUCUUCUCGCCGGUCGCGUUUAAAACCGCAUUUUUUUCGCCUUGCGUUUGGAAACGCGAGCGGUUUUUGCAGCUAACAAAUUAAUUCAUUAAUUCCCAUGCUGACGGGCGUUUUA (((((........((((.((((((((..((.((((............)))).))..)))))))).))))..(((((((.....((((((....)))))).....)))).))))))))... ( -33.80) >DroYak_CAF1 18541 120 + 1 AAUGUAUCUUCUCGCCGGUCGCGUUUAAAACCGCAUUUUUUUCGGCUUGCGAUUGGAAACGCAAGCGGUUUUUGCAGCUACCAAAUUAAUUCAUUAAUUCCAAUGCUGACGGGCGUUUUA ............((((.((((((...((((((((.......(((.....)))..(....)....)))))))))))(((.....((((((....)))))).....)))))).))))..... ( -32.50) >consensus AAUGUAUCUUCUCGCCGGUCGCGUUUAAAACCGCAUUUUUUUCGCCUUGCGUUUGGAAACGCGAGCGGUUUUUGCAGCUACCAAAUUAAUUCAUUAAUUCCAAUGCUGACGGGCGUUUUA ............((((.((((((...((((((((............(((((((....))))))))))))))))))(((.....((((((....)))))).....)))))).))))..... (-31.94 = -32.25 + 0.31)

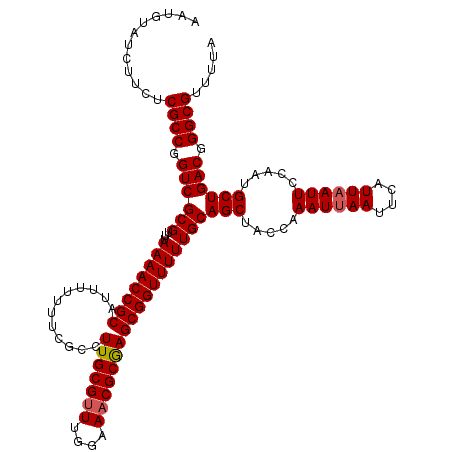
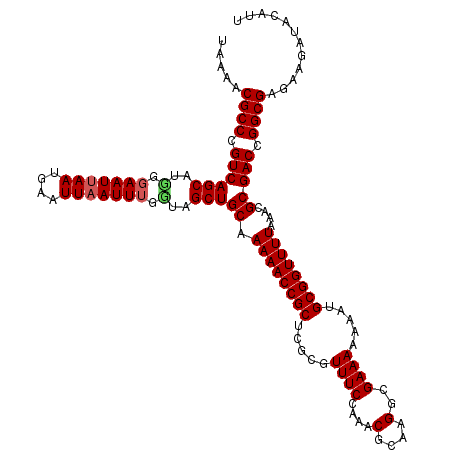
| Location | 9,522,206 – 9,522,326 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -30.41 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9522206 120 - 20766785 UAAAACGCCCGUCAGCAUUGGAAUUAAUGAAUUUAUUAGAUAGCUGCAAAAACCGCUCGCGUUUCCAAACGCAAGGCGAAAAAAAUGCGGUUUUAAACGCGACCGGCGAGAAGAUACAUU .....((((.(((((((((.(((((....)))))....))).)))((.((((((((.....((((....(....)..)))).....))))))))....))))).))))............ ( -31.30) >DroSec_CAF1 18807 120 - 1 UAAAACGCCCGUCAGCAUGGGAAUUAAUGAAUUAAUUUGGUAGCUGCAAAAACCGCUCGCGUUUCCAAACGCAAGGCGAAAAAAAUGCGGUUUUAAACGCGACCGGCGAGAACAUACAUU .....((((.((((((....(((((((....)))))))....)))((.((((((((.....((((....(....)..)))).....))))))))....))))).))))............ ( -32.00) >DroSim_CAF1 19172 120 - 1 UAAAACGCCCGUCAGCAUGGGAAUUAAUGAAUUAAUUUGUUAGCUGCAAAAACCGCUCGCGUUUCCAAACGCAAGGCGAAAAAAAUGCGGUUUUAAACGCGACCGGCGAGAAGAUACAUU .....((((.((((((..(.(((((((....))))))).)..)))((.((((((((.....((((....(....)..)))).....))))))))....))))).))))............ ( -33.20) >DroYak_CAF1 18541 120 - 1 UAAAACGCCCGUCAGCAUUGGAAUUAAUGAAUUAAUUUGGUAGCUGCAAAAACCGCUUGCGUUUCCAAUCGCAAGCCGAAAAAAAUGCGGUUUUAAACGCGACCGGCGAGAAGAUACAUU .....((((.((((((..(.(((((((....))))))).)..))).......(.(((((((........))))))).)........(((........)))))).))))............ ( -33.20) >consensus UAAAACGCCCGUCAGCAUGGGAAUUAAUGAAUUAAUUUGGUAGCUGCAAAAACCGCUCGCGUUUCCAAACGCAAGGCGAAAAAAAUGCGGUUUUAAACGCGACCGGCGAGAAGAUACAUU .....((((.((((((..(.(((((((....))))))).)..)))((.((((((((.....((((....(....)..)))).....))))))))....))))).))))............ (-30.41 = -30.85 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:42 2006