
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,521,614 – 9,522,008 |
| Length | 394 |
| Max. P | 0.999961 |

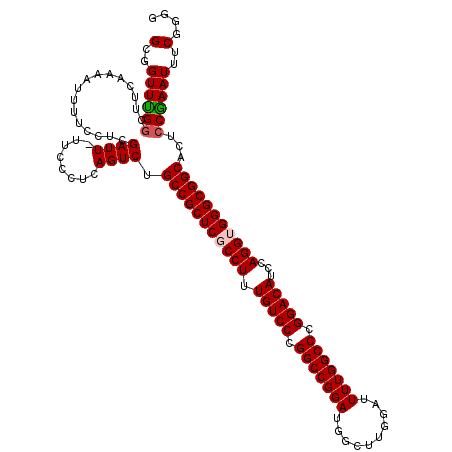
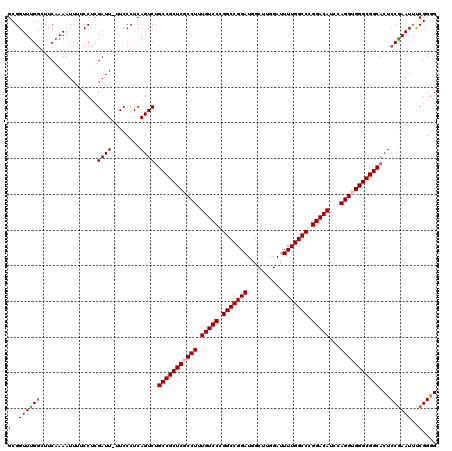
| Location | 9,521,614 – 9,521,734 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -49.52 |
| Consensus MFE | -45.22 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521614 120 + 20766785 GCGGUUUGGAUUCAAAAUUUUCCUCGAUUUUUCCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCAAAUUUCGGGG (..(((((((...............((((........))))((((((((((((.(((((.(((((((..........))))))).)))))...)))))))))))).)))))))..).... ( -50.20) >DroSec_CAF1 18212 119 + 1 GCGGUUCGGCUUCAAAAUUUUCCUCGAUU-UUCCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAAUUCCGGGG .((((((((................((((-.......))))((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))..)))))..)))... ( -50.50) >DroSim_CAF1 18584 119 + 1 GCGGUUUGCCUUCAAAAUUUUCCUCGAUU-UUCCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAAUUCCGGGG ..((....))...............((((-.......))))((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))(((((.....))))) ( -48.80) >DroEre_CAF1 18054 119 + 1 GCGGUUUGGCUUCAAAAUUUUCCUCGAUU-UUCCCUCAGUCAGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGGGGGCGGCACUCCGAAUUUCGGAG ......(((((..((((((......))))-)).....)))))(((((((.(((.(((((.(((((((..........))))))).)))))...))).))))))).(((((.....))))) ( -49.70) >DroYak_CAF1 17958 119 + 1 GCGGUUUGGCUUCAAAAUUUUCCUUGAUU-UUCCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGGGGGCGGCACUCCGAAUUUCGGGG .......((((..((((((......))))-)).....))))((((((((.(((.(((((.(((((((..........))))))).)))))...))).))))))))(((((.....))))) ( -48.40) >consensus GCGGUUUGGCUUCAAAAUUUUCCUCGAUU_UUCCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAAUUUCGGGG (..((((((................((((........)))).(((((((((((.(((((.(((((((..........))))))).)))))...)))))))))))...))))))..).... (-45.22 = -45.50 + 0.28)

| Location | 9,521,614 – 9,521,734 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -42.21 |
| Consensus MFE | -40.03 |
| Energy contribution | -39.75 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521614 120 - 20766785 CCCCGAAAUUUGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGGAAAAAUCGAGGAAAAUUUUGAAUCCAAACCGC ........((((((.((((((.(.(((...(((((.((((................)))).))))).))).).))))))..............((((((....)))))).)))))).... ( -40.39) >DroSec_CAF1 18212 119 - 1 CCCCGGAAUUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGGAA-AAUCGAGGAAAAUUUUGAAGCCGAACCGC ...(((..(((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))((..-..((((((....))))))..))...))). ( -44.89) >DroSim_CAF1 18584 119 - 1 CCCCGGAAUUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGGAA-AAUCGAGGAAAAUUUUGAAGGCAAACCGC ...(((..(((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))....-..((((((....)))))).......))). ( -43.09) >DroEre_CAF1 18054 119 - 1 CUCCGAAAUUCGGAGUGCCGCCCCCCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCUGACUGAGGGAA-AAUCGAGGAAAAUUUUGAAGCCAAACCGC (((((.....))))).(((((.(.(((...(((((.((((................)))).))))).))).).)))))........((..-..((((((....))))))..))....... ( -41.29) >DroYak_CAF1 17958 119 - 1 CCCCGAAAUUCGGAGUGCCGCCCCCCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGGAA-AAUCAAGGAAAAUUUUGAAGCCAAACCGC .(((.....((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))))..-..((((((....))))))........... ( -41.39) >consensus CCCCGAAAUUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGGAA_AAUCGAGGAAAAUUUUGAAGCCAAACCGC .(((.....((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..))))))).....((((((....))))))........... (-40.03 = -39.75 + -0.28)

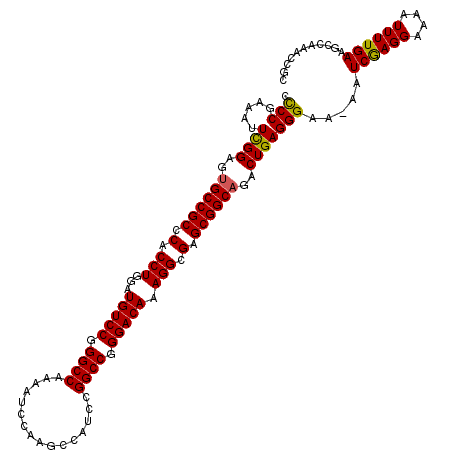
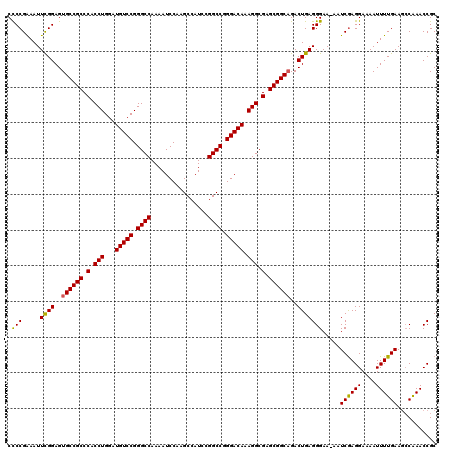
| Location | 9,521,654 – 9,521,774 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -61.94 |
| Consensus MFE | -59.38 |
| Energy contribution | -59.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.91 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521654 120 + 20766785 CUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCAAAUUUCGGGGGCGUGGCUUUUGGGCGGAAAAUUUGUGCGAGCUGUGUGCU .((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))((((.......))))(((..((((..(.(((((...))))).)))))..)))... ( -57.50) >DroSec_CAF1 18251 119 + 1 CUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAAUUCCGGGGGCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGUGCU .((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))(((((.....)))))(((..((((-.(.(((((...))))).)))))..)))... ( -63.20) >DroSim_CAF1 18623 119 + 1 CUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAAUUCCGGGGGCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGCGCU .((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))(((((.....)))))(((..((((-.(.(((((...))))).)))))..)))... ( -65.20) >DroEre_CAF1 18093 119 + 1 CAGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGGGGGCGGCACUCCGAAUUUCGGAGGCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGUGCU ..(((((((.(((.(((((.(((((((..........))))))).)))))...))).))))))).(((((.....)))))(((..((((-.(.(((((...))))).)))))..)))... ( -62.20) >DroYak_CAF1 17997 119 + 1 CUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGGGGGCGGCACUCCGAAUUUCGGGGGCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGUGCU .((((((((.(((.(((((.(((((((..........))))))).)))))...))).))))))))(((((.....)))))(((..((((-.(.(((((...))))).)))))..)))... ( -61.60) >consensus CUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAAUUUCGGGGGCGUGGCUC_UGGGCGGAAAAUUUGUGCGAGCUGUGUGCU ..(((((((((((.(((((.(((((((..........))))))).)))))...))))))))))).(((((.....)))))(((..((((..(.(((((...))))).)))))..)))... (-59.38 = -59.50 + 0.12)

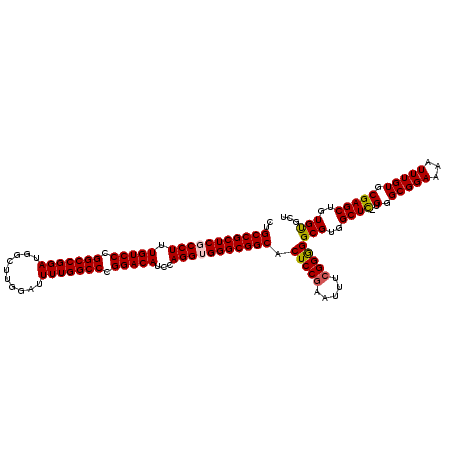
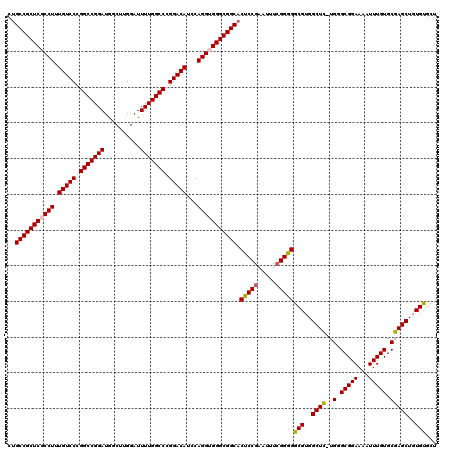
| Location | 9,521,654 – 9,521,774 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -42.89 |
| Consensus MFE | -39.19 |
| Energy contribution | -38.79 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521654 120 - 20766785 AGCACACAGCUCGCACAAAUUUUCCGCCCAAAAGCCACGCCCCCGAAAUUUGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAG ........((((...((((((((..((...........))....))))))))))))(((((.(.(((...(((((.((((................)))).))))).))).).))))).. ( -38.99) >DroSec_CAF1 18251 119 - 1 AGCACACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGCCCCCGGAAUUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAG ........((.(((...........((...-..))..((((((((((((((((.(((.....))))))))).))))))......(((..(((....)))..)))....)))).))))).. ( -42.10) >DroSim_CAF1 18623 119 - 1 AGCGCACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGCCCCCGGAAUUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAG ...((...((((((.............(((-(.((..(((..(((.....))).)))..))....)))).(((((.((((................)))).)))))...)))))).)).. ( -43.39) >DroEre_CAF1 18093 119 - 1 AGCACACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGCCUCCGAAAUUCGGAGUGCCGCCCCCCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCUG (((.....((((((.............(((-(.((...(((((((.....))))).)).))....)))).(((((.((((................)))).)))))...)))))).))). ( -47.09) >DroYak_CAF1 17997 119 - 1 AGCACACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGCCCCCGAAAUUCGGAGUGCCGCCCCCCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAG .((.....((((((.............(((-(.((..(((..(((.....))).)))..))....)))).(((((.((((................)))).)))))...)))))).)).. ( -42.89) >consensus AGCACACAGCUCGCACAAAUUUUCCGCCCA_GAGCCACGCCCCCGAAAUUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAG .((.....((((((.............(((...((..(((..((((...)))).)))..)).....))).(((((.((((................)))).)))))...)))))).)).. (-39.19 = -38.79 + -0.40)


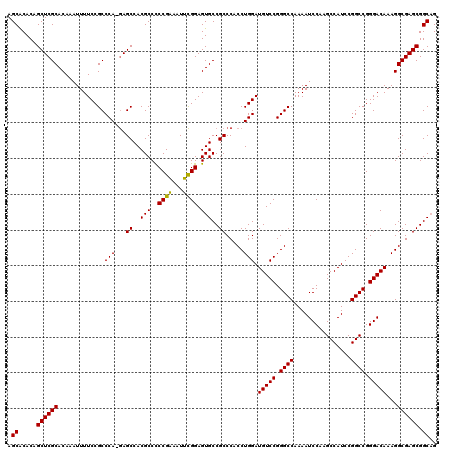
| Location | 9,521,734 – 9,521,854 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -47.64 |
| Consensus MFE | -40.40 |
| Energy contribution | -41.32 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521734 120 + 20766785 GCGUGGCUUUUGGGCGGAAAAUUUGUGCGAGCUGUGUGCUUGCCAGGCUUUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCC ((.((((..((((((((((((((((.((((((.....)))))))))).))))).......(((((..((((..(((((....)).)))..))))..))))))))))))....)))).)). ( -45.90) >DroSec_CAF1 18331 119 + 1 GCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGUGCUUGCCAGGCAUUUCGAUGGUUUUUGGCUCGGAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCC ....(((((-(((((.(((((((((.((((((.....))))))))).(((....))))))))).)))))))..(((((....)).)))...((((....(((((...))))).))))))) ( -46.00) >DroSim_CAF1 18703 119 + 1 GCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGCGCUUGCCAGGCUUUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCGGCCGAGCC ((.((((((-((((((...(((((((((((((((.((....)))))(((((((((.(.(....).))))))))))........))))))...))))))...)))))))...))))).)). ( -49.00) >DroEre_CAF1 18173 119 + 1 GCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGUGCUUGCACGGCUAUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCC (.(..((.(-(((((((((.(.((((((((((.....)))))))))).).))).......(((((..((((..(((((....)).)))..))))..)))))))))))).))..))..... ( -48.90) >DroYak_CAF1 18077 119 + 1 GCGUGGCUC-UGGGCGGAAAAUUUGUGCGAGCUGUGUGCUUGCCGGGCUAUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGGCGAGCC ((.((((.(-((((((...((((((((((((..((.(((((..(((((((............)))))))..))))).))....))))))...))))))...))))))).))))))..... ( -48.40) >consensus GCGUGGCUC_UGGGCGGAAAAUUUGUGCGAGCUGUGUGCUUGCCAGGCUUUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCC ((.((((...(((((((((((((((.((((((.....)))))))))).))))).......(((((..((((..(((((....)).)))..))))..))))))))))).....)))).)). (-40.40 = -41.32 + 0.92)


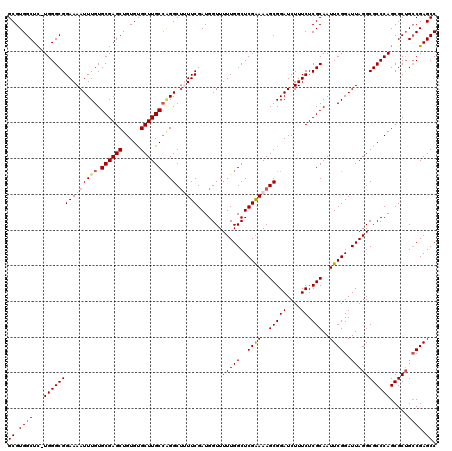
| Location | 9,521,734 – 9,521,854 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -34.20 |
| Energy contribution | -35.40 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521734 120 - 20766785 GGCUCGGCAGCGCUGGGCGCCUAAUCCGAAUUGCGAGAAAGAUCCGCUUUUCGAGCCAAAAACCAUCGAAAAGCCUGGCAAGCACACAGCUCGCACAAAUUUUCCGCCCAAAAGCCACGC (((((((..((((...)))).....))))...(((.(((((....(((((((((...........))))))))).((((.(((.....))).)).))..))))))))......))).... ( -37.10) >DroSec_CAF1 18331 119 - 1 GGCUCGGCAGCGCUGGGCGCCUAAUCCGAAUUGCGAGAAAGAUCCGCUUUCCGAGCCAAAAACCAUCGAAAUGCCUGGCAAGCACACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGC (((((((((.((.(((........((.(((..(((.((....))))).))).))........))).))...))))((((.(((.....))).)).)).............-))))).... ( -35.89) >DroSim_CAF1 18703 119 - 1 GGCUCGGCCGCGCUGGGCGCCUAAUCCGAAUUGCGAGAAAGAUCCGCUUUUCGAGCCAAAAACCAUCGAAAAGCCUGGCAAGCGCACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGC ((((((((...)))(((((...(((......((((((........(((((((((...........)))))))))(((((....)).)))))))))...)))...))))).-))))).... ( -43.10) >DroEre_CAF1 18173 119 - 1 GGCUCGGCAGCGCUGGGCGCCUAAUCCGAAUUGCGAGAAAGAUCCGCUUUUCGAGCCAAAAACCAUCGAAUAGCCGUGCAAGCACACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGC ((((((((.((.....)))))...........(((.(((((....(((.(((((...........))))).))).((((.(((.....))).))))...))))))))...-))))).... ( -41.00) >DroYak_CAF1 18077 119 - 1 GGCUCGCCAGCGCUGGGCGCCUAAUCCGAAUUGCGAGAAAGAUCCGCUUUUCGAGCCAAAAACCAUCGAAUAGCCCGGCAAGCACACAGCUCGCACAAAUUUUCCGCCCA-GAGCCACGC (((((((....))((((((.......((((..(((.((....)))))..))))..............(((.......((.(((.....))).)).......)))))))))-))))).... ( -37.14) >consensus GGCUCGGCAGCGCUGGGCGCCUAAUCCGAAUUGCGAGAAAGAUCCGCUUUUCGAGCCAAAAACCAUCGAAAAGCCUGGCAAGCACACAGCUCGCACAAAUUUUCCGCCCA_GAGCCACGC ((((((((.((.....)))))...........(((.(((((....(((((((((...........)))))))))...((.(((.....))).)).....))))))))....))))).... (-34.20 = -35.40 + 1.20)

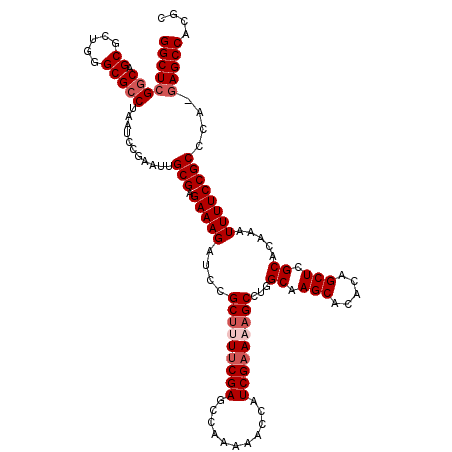

| Location | 9,521,774 – 9,521,894 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -32.90 |
| Energy contribution | -35.42 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521774 120 + 20766785 UGCCAGGCUUUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCCAUAGAGGCAAUAGAGAAAGAGAGCUUUUCCAGGCCAGCCA ...............((((...(((((.(((((((...((((((((.....((((....(((((...))))).))))(((.....)))....))))))))..)))))))..))))))))) ( -48.60) >DroSec_CAF1 18370 120 + 1 UGCCAGGCAUUUCGAUGGUUUUUGGCUCGGAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCCAUAGAGGUAAUAGAGAAAGAUAGCUUUUCCAGGCCAGCCA ...............((((...((((..(((((((..(((((((((.....((((....(((((...))))).))))(((.....)))....))))))))).)).)))))..)))))))) ( -47.60) >DroSim_CAF1 18742 120 + 1 UGCCAGGCUUUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCGGCCGAGCCAUAGAGGCAAUAGAGAAAGAGAGCUUUUCCAGGCCAGCCA ...............((((...(((((.(((((((...((((((((.....((((.....((((...))))..))))(((.....)))....))))))))..)))))))..))))))))) ( -45.70) >DroEre_CAF1 18212 94 + 1 UGCACGGCUAUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCCAUAGAGGC-AUAGAG------------------------- ...........((.(((.(((((((((((((..(((((....)).)))..)))((....(((((...))))).)))))))).)))).)-)).)).------------------------- ( -31.90) >DroYak_CAF1 18116 119 + 1 UGCCGGGCUAUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGGCGAGCCAUAGAGGA-AUAGAGAAAGAGAGCUUUUCCAGGCCAGACA .(((((((((............))))))(((((((...((((((((...((((......(((((...)))))((....))......))-)).))))))))..)))))))..)))...... ( -42.50) >consensus UGCCAGGCUUUUCGAUGGUUUUUGGCUCGAAAAGCGGAUCUUUCUCGCAAUUCGGAUUAGGCGCCCAGCGCUGCCGAGCCAUAGAGGCAAUAGAGAAAGAGAGCUUUUCCAGGCCAGCCA .(((..(((((((((.(.(....).))))))))))...((((((((.....((((....(((((...))))).))))(((.....)))....))))))))...........)))...... (-32.90 = -35.42 + 2.52)

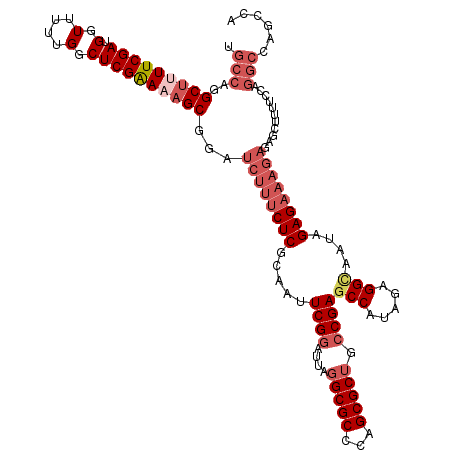
| Location | 9,521,854 – 9,521,971 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -41.68 |
| Consensus MFE | -34.22 |
| Energy contribution | -35.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521854 117 - 20766785 UAACAGGCUGCAAGCAGCCGGAGAAAGCAAAGCAAAGCUGGGACGGAAAAGCCAAGGCGGACUUUCCACUUUU---CGUUUGGCUGGCCUGGAAAAGCUCUCUUUCUCUAUUGCCUCUAU .....(((((....)))))((((((((...(((....(..((.(((..((((.((((.((.....)).)))).---.))))..))).))..)....)))..))))))))........... ( -40.30) >DroSec_CAF1 18450 120 - 1 UAACAGGCUGCAAGCAGCCGGAGAAAGCAAUGCAAAGCUGGGGCGGAAAAGCCAAGGCGCACUUUCCACCUUUCCACGUCUGGCUGGCCUGGAAAAGCUAUCUUUCUCUAUUACCUCUAU .....(((((....)))))((((((((....((...(((..(((......)))..)))............((((((.(((.....))).)))))).))...))))))))........... ( -41.90) >DroSim_CAF1 18822 120 - 1 UAACAGGCUGCAAGCAGCCGGAGAAAGCAAUGCAAAGCUGGGGCGGAAAAUCCAAGGCGCACUUUCCACCUUUCCACGUCUGGCUGGCCUGGAAAAGCUCUCUUUCUCUAUUGCCUCUAU .....(((((....)))))((((...((((((.((((..((((((((((.............)))))...((((((.(((.....))).)))))).)))))))))...)))))))))).. ( -37.42) >DroYak_CAF1 18196 116 - 1 UAACAGGCUGCAAGCAGCCGGAGAAAGCAAAGCAAAGCUGGGGCGGAAAAGCCAAGGUGGACUUUCCACUUUU---CGUUUGUCUGGCCUGGAAAAGCUCUCUUUCUCUAU-UCCUCUAU .....(((((....)))))((((((((...(((....(..((.((((.((((.(((((((.....))))))).---.)))).)))).))..)....)))..))))))))..-........ ( -47.10) >consensus UAACAGGCUGCAAGCAGCCGGAGAAAGCAAAGCAAAGCUGGGGCGGAAAAGCCAAGGCGCACUUUCCACCUUU___CGUCUGGCUGGCCUGGAAAAGCUCUCUUUCUCUAUUGCCUCUAU .....(((((....)))))((((((((....((....(..((.(((..((((.((((.((.....)).)))).....))))..))).))..)....))...))))))))........... (-34.22 = -35.23 + 1.00)


| Location | 9,521,894 – 9,522,008 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -29.89 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9521894 114 - 20766785 ACG---AUGGCGGCGACAAAAAGUCAAUUUUAUCGGAAACUAACAGGCUGCAAGCAGCCGGAGAAAGCAAAGCAAAGCUGGGACGGAAAAGCCAAGGCGGACUUUCCACUUUU---CGUU ...---..(((.(((((.....)))..((((.(((....).....(((((....))))))).)))).....))...)))..((((((((.((....))((.....))..))))---)))) ( -31.00) >DroSec_CAF1 18490 117 - 1 ACG---ACGGCGGCGACAAAAAGUCAAUUUUAUCGGAAACUAACAGGCUGCAAGCAGCCGGAGAAAGCAAUGCAAAGCUGGGGCGGAAAAGCCAAGGCGCACUUUCCACCUUUCCACGUC ..(---(((..(((........))).........(((((......(((((....)))))((.(((((...(((...(((..(((......)))..)))))))))))..))))))).)))) ( -38.30) >DroSim_CAF1 18862 117 - 1 ACG---ACGGCGGCGACAAAAAGUCAAUUUUAUCGGAAACUAACAGGCUGCAAGCAGCCGGAGAAAGCAAUGCAAAGCUGGGGCGGAAAAUCCAAGGCGCACUUUCCACCUUUCCACGUC ..(---(((..(((........))).........(((((......(((((....)))))(((((..((........((....))((.....)).....))..)))))...))))).)))) ( -33.50) >DroYak_CAF1 18235 117 - 1 ACGACGACGGCGGCGACAAAAAGUCAAUUUUAUCGGAAACUAACAGGCUGCAAGCAGCCGGAGAAAGCAAAGCAAAGCUGGGGCGGAAAAGCCAAGGUGGACUUUCCACUUUU---CGUU ..(((((((((.(((((.....)))..((((.(((....).....(((((....))))))).)))).....))...)))).(((......)))(((((((.....))))))))---)))) ( -39.30) >consensus ACG___ACGGCGGCGACAAAAAGUCAAUUUUAUCGGAAACUAACAGGCUGCAAGCAGCCGGAGAAAGCAAAGCAAAGCUGGGGCGGAAAAGCCAAGGCGCACUUUCCACCUUU___CGUC (((....((((.(((((.....)))..((((.(((....).....(((((....))))))).)))).....))...))))(((.(((((.((....))....))))).))).....))). (-29.89 = -29.82 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:38 2006