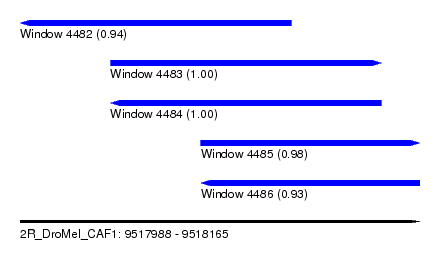
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,517,988 – 9,518,165 |
| Length | 177 |
| Max. P | 0.999624 |

| Location | 9,517,988 – 9,518,108 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9517988 120 - 20766785 AUUUAUUUUGUUUUACGGGCGCGAAAAUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAUUGGAACUAAACGAGAUGUUCGAGUGCCCGGAAAUGCACAC ........(((....(((((((...(((((((((....))))))))).((((((....))))))......(((((((..(((........)))..)))))))))))))).....)))... ( -31.90) >DroSec_CAF1 14979 120 - 1 AUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAUUGGAACUAAACGAGAUGUUCGAGUGCCCGGAAAUGCACAC ........(((....(((((((((..((((((((....))))))))))((((((....))))))......(((((((..(((........)))..)))))))))))))).....)))... ( -30.30) >DroSim_CAF1 15014 120 - 1 AUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAUUGGAACUAAACGAGAUGUUCGAGUGCCCGGAAAUGCACAC ........(((....(((((((((..((((((((....))))))))))((((((....))))))......(((((((..(((........)))..)))))))))))))).....)))... ( -30.30) >DroEre_CAF1 14883 120 - 1 AUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUGUAUUUGAACAAAUUGGAACUAAACGAGAUGUUCGAGUGCCCGGAAAUGCACAC ..................(((((..(((((((((....)))))))))..))).))..((((((((((((((((((((..(((........)))..)))))))))))....)))))).))) ( -31.10) >DroYak_CAF1 14841 120 - 1 AUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAUUGGAACUAAACGAGAUGUUCGAGUGCCCGGAAAUGCACAC ........(((....(((((((((..((((((((....))))))))))((((((....))))))......(((((((..(((........)))..)))))))))))))).....)))... ( -30.30) >consensus AUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAUUGGAACUAAACGAGAUGUUCGAGUGCCCGGAAAUGCACAC ........(((....(((((((((..((((((((....))))))))))((((((....))))))......(((((((..(((........)))..)))))))))))))).....)))... (-30.90 = -30.58 + -0.32)

| Location | 9,518,028 – 9,518,148 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.30 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9518028 120 + 20766785 AUUUGUUCAAAUAUAAACGCCACUCGCAGCAUGAAUUAAUUAACGUUAAUCAAUUUUCGCGCCCGUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUGA .(((((............((.....)).((..(((((.((((....)))).)))))..))((((((((((((.......(((....))).....)))))))))))))))))......... ( -21.30) >DroSec_CAF1 15019 120 + 1 AUUUGUUCAAAUAUAAACGCCACUCGCAGCAUGAAUUAAUUAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUGA ......................(..(((((.(((.(((((....))))))))......(.((((((((((((.......(((....))).....)))))))))))).)..).))))..). ( -24.20) >DroSim_CAF1 15054 120 + 1 AUUUGUUCAAAUAUAAACGCCACUCGCAGCAUGAAUUAAUUAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUGA ......................(..(((((.(((.(((((....))))))))......(.((((((((((((.......(((....))).....)))))))))))).)..).))))..). ( -24.20) >DroEre_CAF1 14923 120 + 1 AUUUGUUCAAAUACAAACGCCACUCGCAGCAUGAAUUAAUUAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACGAGACUGUCUAA .(((((......)))))........((((..(((.(((((....))))))))..(((((.((((((((((((.......(((....))).....)))))))))))).))))))))).... ( -29.70) >DroYak_CAF1 14881 120 + 1 AUUUGUUCAAAUAUAAACGCCACUCGCAGCAUGAAUUAAUUAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUAA .........................(((((.(((.(((((....))))))))......(.((((((((((((.......(((....))).....)))))))))))).)..).)))).... ( -23.70) >consensus AUUUGUUCAAAUAUAAACGCCACUCGCAGCAUGAAUUAAUUAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUGA .(((((......)))))........(((((.(((.(((((....))))))))......(.((((((((((((.......(((....))).....)))))))))))).)..).)))).... (-23.62 = -23.30 + -0.32)

| Location | 9,518,028 – 9,518,148 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -28.92 |
| Energy contribution | -28.64 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9518028 120 - 20766785 UCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUACGGGCGCGAAAAUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAU .....((.((((((((((..((((((.....(((....))).......))))))..)))))))).(((((((((....)))))))))........)).))..(((((....))))).... ( -26.10) >DroSec_CAF1 15019 120 - 1 UCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAU .....((.(((((((((((.((((((.....(((....))).......)))))).))))))))).(((((((((....)))))))))........)).))..(((((....))))).... ( -28.90) >DroSim_CAF1 15054 120 - 1 UCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAU .....((.(((((((((((.((((((.....(((....))).......)))))).))))))))).(((((((((....)))))))))........)).))..(((((....))))).... ( -28.90) >DroEre_CAF1 14923 120 - 1 UUAGACAGUCUCGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUGUAUUUGAACAAAU .....((.(((((((((((.((((((.....(((....))).......)))))).))))))))).(((((((((....)))))))))........)).))..((((((......)))))) ( -30.50) >DroYak_CAF1 14881 120 - 1 UUAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAU .....((.(((((((((((.((((((.....(((....))).......)))))).))))))))).(((((((((....)))))))))........)).))..(((((....))))).... ( -28.90) >consensus UCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUAAUUAAUUCAUGCUGCGAGUGGCGUUUAUAUUUGAACAAAU .....((.(((((((((((.((((((.....(((....))).......)))))).))))))))).(((((((((....)))))))))........)).))..(((((....))))).... (-28.92 = -28.64 + -0.28)

| Location | 9,518,068 – 9,518,165 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -21.04 |
| Energy contribution | -20.85 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9518068 97 + 20766785 UAACGUUAAUCAAUUUUCGCGCCCGUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUGAAUGGCAUUUAGUACAUG--- ....((((.(((((..((..((((((((((((.......(((....))).....))))))))))))....))..)).))).))))............--- ( -19.20) >DroSec_CAF1 15059 100 + 1 UAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUGAAUGGCAUUUAGUGAAUGGAA .........((....(((((((((((((((((.......(((....))).....)))))))))))).......((((.....))))....)))))..)). ( -26.70) >DroEre_CAF1 14963 99 + 1 UAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACGAGACUGUCUAAAUGGCAUUUCGUACUUGGA- .........((((.......((((((((((((.......(((....))).....))))))))))))((((((.((((.....))))))))))..)))).- ( -28.20) >DroYak_CAF1 14921 100 + 1 UAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUAAAUGGCAUUUCGUACUUAGAA ..(((.............(.((((((((((((.......(((....))).....)))))))))))).).....((((.....))))...)))........ ( -22.70) >consensus UAACGUUAAUCAACUUUCGCGCCCAUAAAACAAAAUAAAUAGAAAACUAGAAAAUGUUUUGUGGGCACAAGACUGUCUAAAUGGCAUUUAGUACAUGGA_ ....................((((((((((((.......(((....))).....))))))))))))((.(((.((((.....))))))).))........ (-21.04 = -20.85 + -0.19)

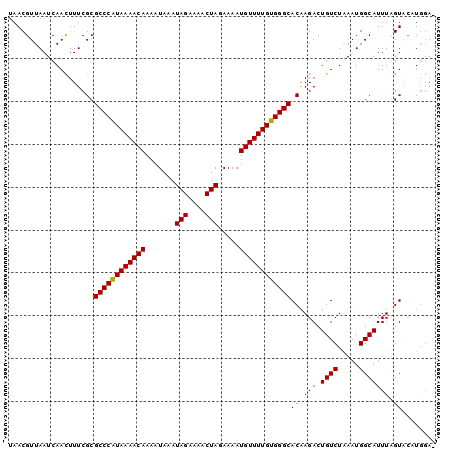
| Location | 9,518,068 – 9,518,165 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.33 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
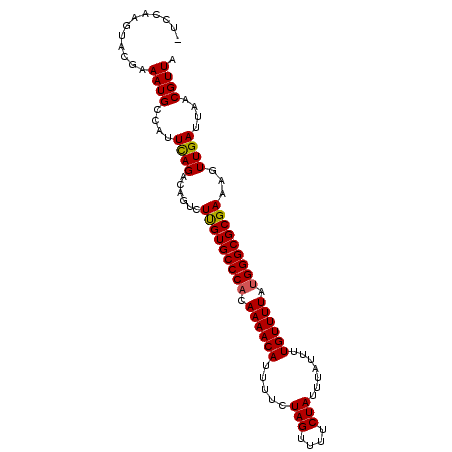
>2R_DroMel_CAF1 9518068 97 - 20766785 ---CAUGUACUAAAUGCCAUUCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUACGGGCGCGAAAAUUGAUUAACGUUA ---......................((((.((((((((..((((((.....(((....))).......))))))..))))))))..)))).......... ( -18.00) >DroSec_CAF1 15059 100 - 1 UUCCAUUCACUAAAUGCCAUUCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUA ............((((....((((......(((((((((.((((((.....(((....))).......)))))).)))))))))...))))....)))). ( -21.70) >DroEre_CAF1 14963 99 - 1 -UCCAAGUACGAAAUGCCAUUUAGACAGUCUCGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUA -......................(((....(((((((((.((((((.....(((....))).......)))))).)))))))))..(((....)))))). ( -22.40) >DroYak_CAF1 14921 100 - 1 UUCUAAGUACGAAAUGCCAUUUAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUA .(((((((..........)))))))(((..(((((((((.((((((.....(((....))).......)))))).))))))).))..))).......... ( -22.20) >consensus _UCCAAGUACGAAAUGCCAUUCAGACAGUCUUGUGCCCACAAAACAUUUUCUAGUUUUCUAUUUAUUUUGUUUUAUGGGCGCGAAAGUUGAUUAACGUUA ............((((....((((......(((((((((.((((((.....(((....))).......)))))).)))))))))...))))....)))). (-20.51 = -20.33 + -0.19)

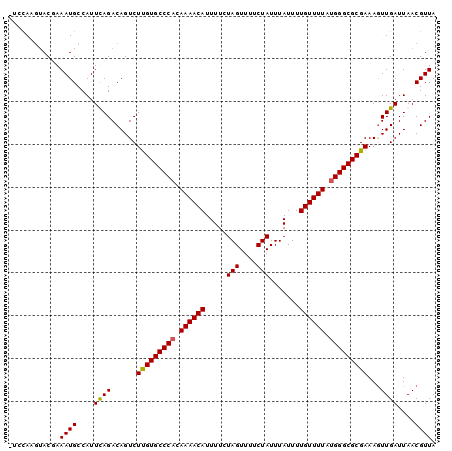
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:24 2006