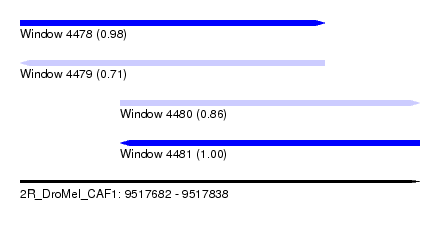
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,517,682 – 9,517,838 |
| Length | 156 |
| Max. P | 0.999780 |

| Location | 9,517,682 – 9,517,801 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9517682 119 + 20766785 UACUAAGUAACUGAGGAAAAUGGAGAAAA-AAAAAUACUAGAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAG ..((..((((((.................-.......(((......)))........((((((((((.......(((.(((.....))).)))....))))))))))))))))..))... ( -23.90) >DroSec_CAF1 14676 120 + 1 UACUAAGUAACUGAGGAAAAUAGAGAAAGAAGAAGUACUAGAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAG ..((..((((((.........................(((......)))........((((((((((.......(((.(((.....))).)))....))))))))))))))))..))... ( -23.90) >DroSim_CAF1 14697 120 + 1 UACUAAGUAACUGAGGAAAAUAGAGAAAGAAAAAAUACUAGAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAG ..((..((((((.........................(((......)))........((((((((((.......(((.(((.....))).)))....))))))))))))))))..))... ( -23.90) >DroYak_CAF1 14539 118 + 1 UACUAAGUAACCGAGGAAAAUAGAGAAAAGAAAAAUACUAGAAAAAUG--GGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAG ..((..((((((.........................(((......))--)......((((((((((.......(((.(((.....))).)))....))))))))))))))))..))... ( -26.50) >consensus UACUAAGUAACUGAGGAAAAUAGAGAAAAAAAAAAUACUAGAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAG ..((..((((((.........................(((......)))........((((((((((.......(((.(((.....))).)))....))))))))))))))))..))... (-23.39 = -23.45 + 0.06)

| Location | 9,517,682 – 9,517,801 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -17.52 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9517682 119 - 20766785 CUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUCUAGUAUUUUU-UUUUCUCCAUUUUCCUCAGUUACUUAGUA .....(((.....((((((((((.((..((((.............))))..))))))))))))........)))................-............................. ( -16.74) >DroSec_CAF1 14676 120 - 1 CUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUCUAGUACUUCUUCUUUCUCUAUUUUCCUCAGUUACUUAGUA .....(((.....((((((((((.((..((((.............))))..))))))))))))........))).............................................. ( -16.74) >DroSim_CAF1 14697 120 - 1 CUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUCUAGUAUUUUUUCUUUCUCUAUUUUCCUCAGUUACUUAGUA .....(((.....((((((((((.((..((((.............))))..))))))))))))........))).............................................. ( -16.74) >DroYak_CAF1 14539 118 - 1 CUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCC--CAUUUUUCUAGUAUUUUUCUUUUCUCUAUUUUCCUCGGUUACUUAGUA ...((.((.((((((((((((((.((..((((.............))))..)))))))))))).......--........(((..............)))........)))).)).)).. ( -19.86) >consensus CUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUCUAGUAUUUUUUCUUUCUCUAUUUUCCUCAGUUACUUAGUA ......(....).((((((((((.((..((((.............))))..))))))))))))......................................................... (-16.32 = -16.32 + 0.00)


| Location | 9,517,721 – 9,517,838 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9517721 117 + 20766785 GAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAGA---CAGAGAUGGAAAAUGCAGAGCGGGAGGGCGAUAUGA .................((((((((((.......(((.(((.....))).)))....)))))))))).((((((.......---((....)).....(((...)))....)))))).... ( -24.50) >DroSec_CAF1 14716 117 + 1 GAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAGA---CUGAGAUGGGAAGUGCAGAGCGGGAAGGCGAUAUGA .......(((((((.....)))))))....((((((((((((.(((((....((((...(.....).)))).(((......---......)))...))))).)))..))))))))).... ( -28.30) >DroSim_CAF1 14737 117 + 1 GAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAGA---CAGAGAUGGGAAGUGCAGAGCGGGAGGGCGAUAUGA .......(((((((.....)))))))....((((((((((((.(((((....((((...(.....).)))).(((......---......)))...))))).)))..))))))))).... ( -27.90) >DroEre_CAF1 14643 112 + 1 ----GAUG-AUGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAGA---AAGAGAUGGGAAAUGCUGAGCGGGAGGGCGAUAUGA ----....-........((((((((((.......(((.(((.....))).)))....)))))))))).((((((.....(.---.((..((.....)).))...).....)))))).... ( -24.60) >DroYak_CAF1 14579 118 + 1 GAAAAAUG--GGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAGAAAAAAGAGAUGGGAAAUGGUGAGCGGGAGGGCGAUAUGA ......((--(.((...((((((((((.......(((.(((.....))).)))....))))))))))..)).)))............................((......))....... ( -23.90) >consensus GAAAAAUGGGAGAGAAAUGUUUUCCCUGAAAUUGCUUUUGUUGUGUACACAAGACCAAGGGAAAACAGGUUGCCAAGAAGA___CAGAGAUGGGAAAUGCAGAGCGGGAGGGCGAUAUGA .................((((((((((.......(((.(((.....))).)))....)))))))))).((((((....................................)))))).... (-21.84 = -21.84 + -0.00)

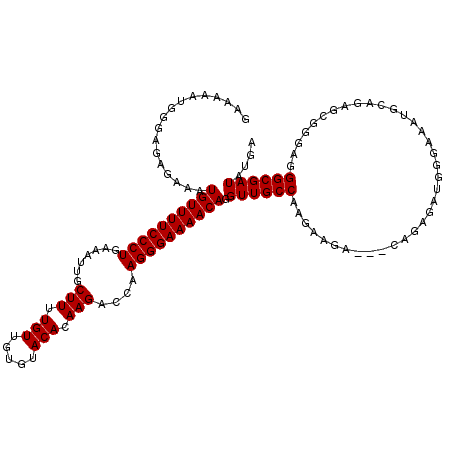
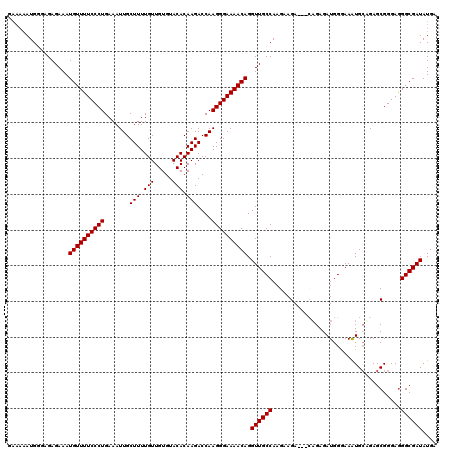
| Location | 9,517,721 – 9,517,838 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9517721 117 - 20766785 UCAUAUCGCCCUCCCGCUCUGCAUUUUCCAUCUCUG---UCUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUC .......((......)).................((---((.....))))...((((((((((.((..((((.............))))..))))))))))))................. ( -19.52) >DroSec_CAF1 14716 117 - 1 UCAUAUCGCCUUCCCGCUCUGCACUUCCCAUCUCAG---UCUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUC .......(((........(((............)))---.......)))....((((((((((.((..((((.............))))..))))))))))))................. ( -20.88) >DroSim_CAF1 14737 117 - 1 UCAUAUCGCCCUCCCGCUCUGCACUUCCCAUCUCUG---UCUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUC .......((......)).................((---((.....))))...((((((((((.((..((((.............))))..))))))))))))................. ( -19.52) >DroEre_CAF1 14643 112 - 1 UCAUAUCGCCCUCCCGCUCAGCAUUUCCCAUCUCUU---UCUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCAU-CAUC---- .......(((..........................---.......)))....((((((((((.((..((((.............))))..))))))))))))........-....---- ( -17.93) >DroYak_CAF1 14579 118 - 1 UCAUAUCGCCCUCCCGCUCACCAUUUCCCAUCUCUUUUUUCUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCC--CAUUUUUC .......((......))............................(((.....((((((((((.((..((((.............))))..))))))))))))......)--))...... ( -18.32) >consensus UCAUAUCGCCCUCCCGCUCUGCAUUUCCCAUCUCUG___UCUUCUUGGCAACCUGUUUUCCCUUGGUCUUGUGUACACAACAAAAGCAAUUUCAGGGAAAACAUUUCUCUCCCAUUUUUC .......(((....................................)))....((((((((((.((..((((.............))))..))))))))))))................. (-18.08 = -18.08 + 0.00)

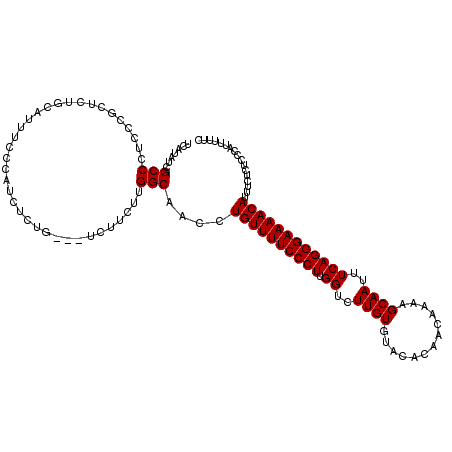
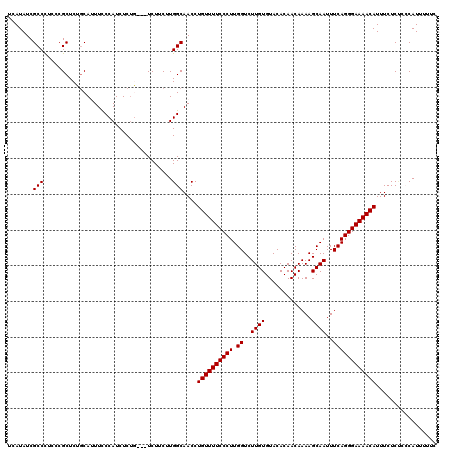
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:19 2006