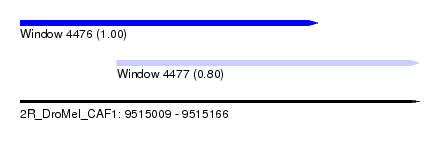
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,515,009 – 9,515,166 |
| Length | 157 |
| Max. P | 0.997420 |

| Location | 9,515,009 – 9,515,126 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.68 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -28.22 |
| Energy contribution | -27.82 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9515009 117 + 20766785 CGAAAA--UCAAAUUGCCUGGAAAAAGGGUGAAAAAGGUGGGGAAUAUGUUGAAUAUGAAAUU-UCUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUUAC .(((.(--((((((......(((((..(((.(((.((((((((.((((.....))))....((-(((....)))))...))))))))...))).)))..)))))))))))).)))..... ( -25.70) >DroSec_CAF1 12018 117 + 1 CGAAAA--UGAAAUUGCCUGGAAAAAGGGUGAAAAAGGUGGGGAAUAUGUUGAAUAUGAAAUU-GUUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUCAC ......--.....(..(((.......)))..).....((((((((((..(..((((.(((...-((((((((((..((......))))))))))))...))).))))..))))))))))) ( -30.20) >DroSim_CAF1 12059 119 + 1 CGAAAAAUUCAAAUUGCCUGGAAAAAGGGUGAAAAAGGUGGGGAAUAUGUUGAAUAUGAAAUU-GUUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUUAC .............(..(((.......)))..).....((((((((((..(..((((.(((...-((((((((((..((......))))))))))))...))).))))..))))))))))) ( -28.10) >DroEre_CAF1 12294 118 + 1 CGAAAA--UCAAAUUGCCUGGAAAAAGGGUGAAAAAGGUGGAGAAUAUGUUGAAUAUGAAAUUUGCUAGACUGAAAGAGCCCUAUCUCUGUUUGGCUUGUUUUUGUUUGGUAUUCUUUAC ......--.....(..(((.......)))..).....((((((((((..(..((((.(((((..(((((((....((((......)))))))))))..)))))))))..))))))))))) ( -29.20) >DroYak_CAF1 12153 118 + 1 CAAAAA--UCAAAUUGCCUGGAAAAAGGGUGAAAAAGGUGGAGAAUAUGUUGAAUAUGAAAUUUGCUAGGCAGAAGGAGCCCUAUCUCUGUUUGGCUUGUUUUUGUUUGGUAUUCUUUAC ......--.....(..(((.......)))..).....((((((((((..(..((((.(((((..(((((((((((((...)))...))))))))))..)))))))))..))))))))))) ( -33.20) >consensus CGAAAA__UCAAAUUGCCUGGAAAAAGGGUGAAAAAGGUGGGGAAUAUGUUGAAUAUGAAAUU_GCUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUUAC .............(..(((.......)))..).....((((((((((..(..((((.(((((..((((((((((..((......))))))))))))..)))))))))..))))))))))) (-28.22 = -27.82 + -0.40)

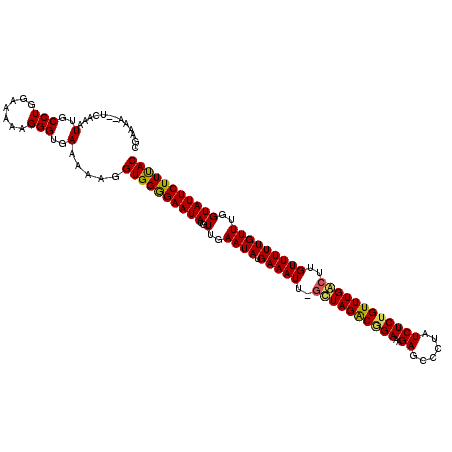
| Location | 9,515,047 – 9,515,166 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.31 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9515047 119 + 20766785 GGGAAUAUGUUGAAUAUGAAAUU-UCUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUUACUUGAUUUCAUACCCGUCACACGAACAUUACGUAGGGUAUA .(((((..((.((((((.((((.-.(.((.((((.((((......)))).)))).)).).....)))).))))))...))...)))))((((((.....(((.......))).)))))). ( -24.50) >DroSec_CAF1 12056 119 + 1 GGGAAUAUGUUGAAUAUGAAAUU-GUUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUCACUUGAUUUUAUACCCGUCACACGAACAUUACGUAGGGUAUA .(((..(((..(((((.(((...-((((((((((..((......))))))))))))...))).)))))..)))..))).........(((((((.....(((.......))).))))))) ( -27.90) >DroSim_CAF1 12099 119 + 1 GGGAAUAUGUUGAAUAUGAAAUU-GUUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUUACUUGAUUUUAUACCCGUCACACGAACAUUACGUAGGGUAUA (((((((..(..((((.(((...-((((((((((..((......))))))))))))...))).))))..))))))))..........(((((((.....(((.......))).))))))) ( -27.60) >DroEre_CAF1 12332 120 + 1 GAGAAUAUGUUGAAUAUGAAAUUUGCUAGACUGAAAGAGCCCUAUCUCUGUUUGGCUUGUUUUUGUUUGGUAUUCUUUACUUGAUUUUAUACCCGGCACACGAACAUUACGAAGGGUAUA (((((((..(..((((.(((((..(((((((....((((......)))))))))))..)))))))))..))))))))..........(((((((..(.............)..))))))) ( -27.72) >DroYak_CAF1 12191 120 + 1 GAGAAUAUGUUGAAUAUGAAAUUUGCUAGGCAGAAGGAGCCCUAUCUCUGUUUGGCUUGUUUUUGUUUGGUAUUCUUUACUUGAUUUUAUACCCUGCACACCGAGAUUACGUAGGGUAUA (((((((..(..((((.(((((..(((((((((((((...)))...))))))))))..)))))))))..))))))))..........((((((((((.............)))))))))) ( -36.82) >consensus GGGAAUAUGUUGAAUAUGAAAUU_GCUAGACGGAAAGAGCCCUAUCUCUGUUUGACUUGUUUUUGUUUGGUAUUCUUUACUUGAUUUUAUACCCGUCACACGAACAUUACGUAGGGUAUA (((((((..(..((((.(((((..((((((((((..((......))))))))))))..)))))))))..))))))))..........(((((((...................))))))) (-25.35 = -25.31 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:16 2006