
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,510,576 – 9,510,798 |
| Length | 222 |
| Max. P | 0.999484 |

| Location | 9,510,576 – 9,510,696 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -38.90 |
| Energy contribution | -39.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
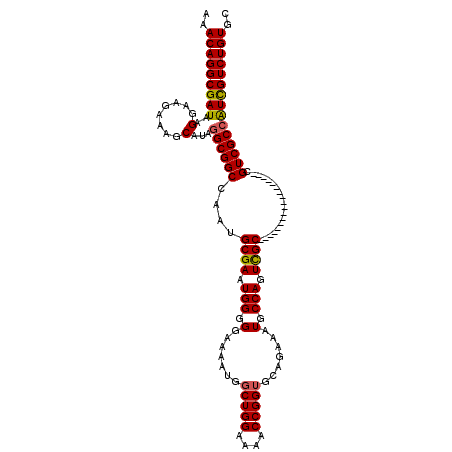
>2R_DroMel_CAF1 9510576 120 + 20766785 CGCUGCAUUGUAGCAGGCGCUCAGCUGCAUUCACCGCACGAUUGUGUGCAGUGGCCGCAUGUUGCAAGUUGUUGCACCAGCAACACUCAAGCGAAUCUUUCGCGAAAGCCCGAGACGCAG ((((...((((((((.(((..((.((((((.((.........)).))))))))..))).))))))))..((((((....))))))....))))..((((..((....))..))))..... ( -42.50) >DroSec_CAF1 7600 118 + 1 CGCUGCAUUGUAGCAGGCGCUCAGCUGCAUUCACCGCACGAUUG--UGCAGUGGCCGCAUGUUGCAAGUUGUUGCACCAGCAACACUCAAGCGAAUCUUUCGCGAAAGCCCGAGACGCAG ((((...((((((((.(((..((.((((((((.......))..)--)))))))..))).))))))))..((((((....))))))....))))..((((..((....))..))))..... ( -41.90) >DroSim_CAF1 7623 118 + 1 CGCUGCAUUGUAGCAGGCGCUCAGCUGCAUUCACCGCACGAUUG--UGCAGUGGCCGCAUGUUGCAAGUUGUUGCACCAGCAACACUCAAGCGAAUCUUUCGCGAAAGCCCGAGACGCAG ((((...((((((((.(((..((.((((((((.......))..)--)))))))..))).))))))))..((((((....))))))....))))..((((..((....))..))))..... ( -41.90) >DroEre_CAF1 7726 118 + 1 CGCUGCAUUGUAGCAGGCGCUCAGCUGCAUUAACCGCACGAUUG--UGCAGUGGUCGCAUGUUGCAAGUUGUUGCACCAGCAACACUCCAGCGAAUCUUUCGCGAAAGCCCGAGACGCAG (((((..((((((((.(((..((.((((((.............)--)))))))..))).))))))))..((((((....))))))...)))))..((((..((....))..))))..... ( -44.02) >DroYak_CAF1 7632 118 + 1 CGCUGCAUUGUAGCAGGCGCUCAGCUGCAUUCACCGCACGAUUG--UGCAGUGGUCGCAUGUGGCAAGUUGUUGCACCAGCAACACUCCAGCGAAUCUUUCGCGAAAGCACGAGACGCAG .(((((...)))))..((((((.(((((..((((.((((....)--))).))))..)))..(((..(((.(((((....))))))))))))).........((....))..))).))).. ( -44.10) >consensus CGCUGCAUUGUAGCAGGCGCUCAGCUGCAUUCACCGCACGAUUG__UGCAGUGGCCGCAUGUUGCAAGUUGUUGCACCAGCAACACUCAAGCGAAUCUUUCGCGAAAGCCCGAGACGCAG ((((...((((((((.(((..((.(((((.....(....)......)))))))..))).))))))))..((((((....))))))....))))..((((..((....))..))))..... (-38.90 = -39.10 + 0.20)

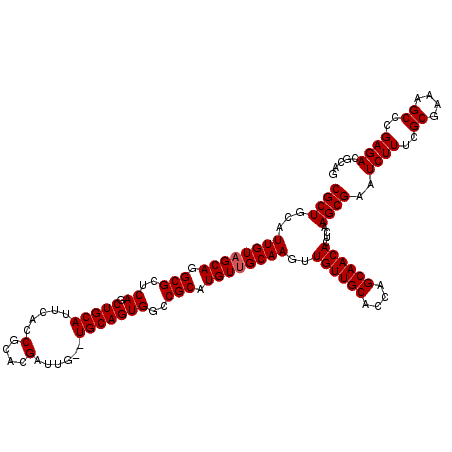
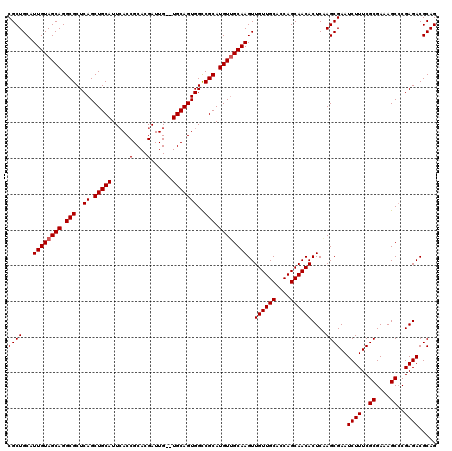
| Location | 9,510,576 – 9,510,696 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -49.92 |
| Consensus MFE | -45.28 |
| Energy contribution | -45.88 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510576 120 - 20766785 CUGCGUCUCGGGCUUUCGCGAAAGAUUCGCUUGAGUGUUGCUGGUGCAACAACUUGCAACAUGCGGCCACUGCACACAAUCGUGCGGUGAAUGCAGCUGAGCGCCUGCUACAAUGCAGCG ..(((.(((((((((..((((.....))))..))))(((((.(((......))).))))).((((..((((((((......))))))))..)))).))))))))((((......)))).. ( -51.80) >DroSec_CAF1 7600 118 - 1 CUGCGUCUCGGGCUUUCGCGAAAGAUUCGCUUGAGUGUUGCUGGUGCAACAACUUGCAACAUGCGGCCACUGCA--CAAUCGUGCGGUGAAUGCAGCUGAGCGCCUGCUACAAUGCAGCG ..(((.(((((((((..((((.....))))..))))(((((.(((......))).))))).((((..(((((((--(....))))))))..)))).))))))))((((......)))).. ( -51.60) >DroSim_CAF1 7623 118 - 1 CUGCGUCUCGGGCUUUCGCGAAAGAUUCGCUUGAGUGUUGCUGGUGCAACAACUUGCAACAUGCGGCCACUGCA--CAAUCGUGCGGUGAAUGCAGCUGAGCGCCUGCUACAAUGCAGCG ..(((.(((((((((..((((.....))))..))))(((((.(((......))).))))).((((..(((((((--(....))))))))..)))).))))))))((((......)))).. ( -51.60) >DroEre_CAF1 7726 118 - 1 CUGCGUCUCGGGCUUUCGCGAAAGAUUCGCUGGAGUGUUGCUGGUGCAACAACUUGCAACAUGCGACCACUGCA--CAAUCGUGCGGUUAAUGCAGCUGAGCGCCUGCUACAAUGCAGCG ..(((.(((((((((..((((.....))))..))))(((((.(((......))).))))).((((...((((((--(....)))))))...)))).))))))))((((......)))).. ( -47.60) >DroYak_CAF1 7632 118 - 1 CUGCGUCUCGUGCUUUCGCGAAAGAUUCGCUGGAGUGUUGCUGGUGCAACAACUUGCCACAUGCGACCACUGCA--CAAUCGUGCGGUGAAUGCAGCUGAGCGCCUGCUACAAUGCAGCG ..(((.((((.((....((((.....))))(((..((((((....)))))).....)))..((((..(((((((--(....))))))))..)))))))))))))((((......)))).. ( -47.00) >consensus CUGCGUCUCGGGCUUUCGCGAAAGAUUCGCUUGAGUGUUGCUGGUGCAACAACUUGCAACAUGCGGCCACUGCA__CAAUCGUGCGGUGAAUGCAGCUGAGCGCCUGCUACAAUGCAGCG ..(((.(((((((((..((((.....))))..))))(((((.(((......))).))))).((((..(((((((........)))))))..)))).))))))))((((......)))).. (-45.28 = -45.88 + 0.60)

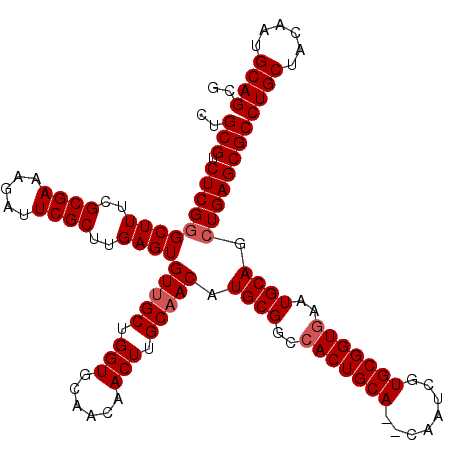
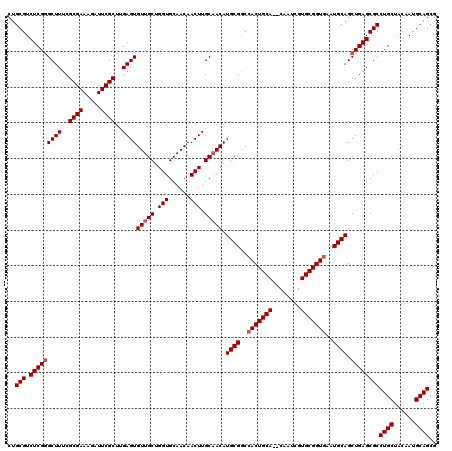
| Location | 9,510,696 – 9,510,798 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -31.18 |
| Energy contribution | -30.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510696 102 - 20766785 AAACAGGCGAUAAGGAAGAAAGCAUAGGCGGCCAAUGCGAAUGGGGGAAAAUGGCUGGAAAACCGGUGCAGAAAUGCCAGUCGC------------------CGUCGCCAUCGUCUGUGC ..(((((((((..(........)...((((((....((((.(((.(.....(((((((....))))).))....).))).))))------------------.))))))))))))))).. ( -37.50) >DroSec_CAF1 7718 102 - 1 AAACAGGCGAUAAGGAAGAAAGCAUAGGCGGCCAAUGCGAAUGGGGGAAAAUGGCUGGAAAACCGGUGCAGAAAUGCCAGUUGC------------------CGUCGCCAUCGUCUGUGC ..(((((((((..(........)...((((((....((((.(((.(.....(((((((....))))).))....).))).))))------------------.))))))))))))))).. ( -35.40) >DroSim_CAF1 7741 102 - 1 AAACAGGCGAUAAGGAAGAAAGCAUAGGCGGCCAAUGCGAAUGGGGGAAAAUGGCUGGAAAACCGGUGCAGAAAUGCCAGUUGC------------------CGUCGCCAUCGUCUGUGC ..(((((((((..(........)...((((((....((((.(((.(.....(((((((....))))).))....).))).))))------------------.))))))))))))))).. ( -35.40) >DroEre_CAF1 7844 120 - 1 AAACAGGCGAUAAGGAAGAAAGCAUAGGCGGCCAAUGCGAAUGGGGGAAAAUGCCUGGAAAACCGGUGCAGAAAUGCCAGUCGCCCUCGCCCUCGCCGUCGCCGUCGCCGUUGUCUGUGU ..(((((((((..((..((..((...((((((....((((..(((.((...((.((((....)))).(((....))))).)).)))))))....))))))))..)).))))))))))).. ( -44.90) >DroYak_CAF1 7750 117 - 1 AAACAGGCGAUAAGGAAGAAAGCAUAGGCGGCCAACGCGAAUGGGGGAAAAUAGCUGGAAAACCGGUGCAGAAAUGCCAGUCGCCGACG---GCGAGGGGGACGUCGCAGUCGUCUGUGU ..(((((((((...............((((((.....................(((((....)))))(((....)))..))))))....---((((.(....).)))).))))))))).. ( -44.20) >consensus AAACAGGCGAUAAGGAAGAAAGCAUAGGCGGCCAAUGCGAAUGGGGGAAAAUGGCUGGAAAACCGGUGCAGAAAUGCCAGUCGC__________________CGUCGCCAUCGUCUGUGC ..(((((((((..(........)...((((((....((((.(((.(.......(((((....))))).......).))).))))...................))))))))))))))).. (-31.18 = -30.94 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:56 2006