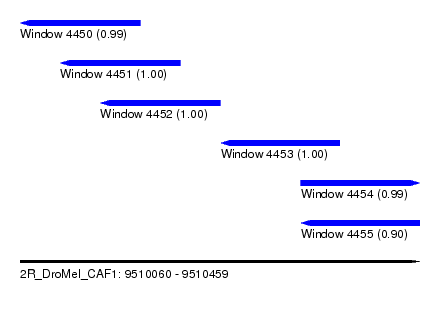
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,510,060 – 9,510,459 |
| Length | 399 |
| Max. P | 0.999919 |

| Location | 9,510,060 – 9,510,180 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510060 120 - 20766785 GAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGGACGGCAUUAGAGCGUCGAACAGAGAUGGCAGACGCACAGA ..(((((((.((....(.(((((((((.....)))).))))))....)).)))))))........(((((............)))))....(((((...((....))...)))))..... ( -32.70) >DroSec_CAF1 7084 120 - 1 GAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGGACGGCAACAGAGCGUCGGACAGAGAUGGCAGACGCACAGA ..(((((((.((....(.(((((((((.....)))).))))))....)).))))))).........((((............)))).....(((((...((....))...)))))..... ( -32.40) >DroSim_CAF1 7109 120 - 1 GAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGGACGGCAACAGAGCGUCGGACAGAGAUGGCAGACGCACAGA ..(((((((.((....(.(((((((((.....)))).))))))....)).))))))).........((((............)))).....(((((...((....))...)))))..... ( -32.40) >DroEre_CAF1 7240 120 - 1 GAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGGAUGGCAACAGAGCGUCGGACAGAGAUGGGAGACGCACAGA ..(((((((.((....(.(((((((((.....)))).))))))....)).))))))).........((((((.......).))))).....(((((...((....))...)))))..... ( -33.60) >DroYak_CAF1 7119 120 - 1 GAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGGAUGGCAACAGAGCGUCGGACAGAGACGGCAGACGCACAGA ..(((((((.((....(.(((((((((.....)))).))))))....)).))))))).........((((((.......).))))).....(((((...(......)...)))))..... ( -32.10) >consensus GAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGGACGGCAACAGAGCGUCGGACAGAGAUGGCAGACGCACAGA ..(((((((.((....(.(((((((((.....)))).))))))....)).))))))).........((((............)))).....(((((...((....))...)))))..... (-31.64 = -31.84 + 0.20)

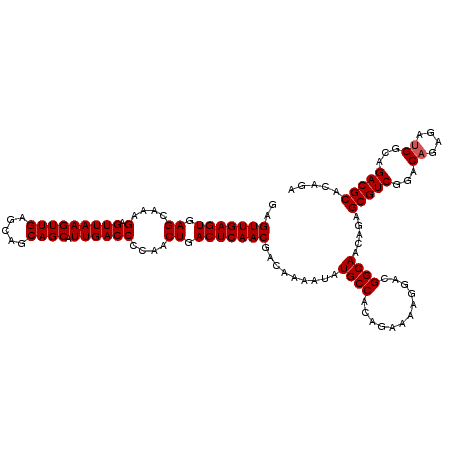
| Location | 9,510,100 – 9,510,220 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510100 120 - 20766785 UCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGG ...((((((((((....))))))))))....(.(((.(.((.(((((((.((....(.(((((((((.....)))).))))))....)).)))))))..))...).)))).......... ( -34.60) >DroSec_CAF1 7124 120 - 1 UCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGG ...((((((((((....))))))))))....(.(((.(.((.(((((((.((....(.(((((((((.....)))).))))))....)).)))))))..))...).)))).......... ( -34.60) >DroSim_CAF1 7149 120 - 1 UCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGG ...((((((((((....))))))))))....(.(((.(.((.(((((((.((....(.(((((((((.....)))).))))))....)).)))))))..))...).)))).......... ( -34.60) >DroEre_CAF1 7280 120 - 1 UCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGG ...((((((((((....))))))))))....(.(((.(.((.(((((((.((....(.(((((((((.....)))).))))))....)).)))))))..))...).)))).......... ( -34.60) >DroYak_CAF1 7159 120 - 1 UCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGG ...((((((((((....))))))))))....(.(((.(.((.(((((((.((....(.(((((((((.....)))).))))))....)).)))))))..))...).)))).......... ( -34.60) >consensus UCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUGACCCCAACUGACUCAACGACAAAAUAUGCCACAGAAAAGG ...((((((((((....))))))))))....(.(((.(.((.(((((((.((....(.(((((((((.....)))).))))))....)).)))))))..))...).)))).......... (-34.60 = -34.60 + 0.00)


| Location | 9,510,140 – 9,510,260 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510140 120 - 20766785 ACAGGAGAAAAACCCGAAAAAACAAGCGUACAACAAAAGCUCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUG ...((........))..........((((.((((...(((((.((((((((((....))))))))))..(((..(((......)))..))).....)))))..)))).)).))....... ( -30.40) >DroSec_CAF1 7164 120 - 1 ACAGGAGAAAAACCCGAAAAAAAAAGCGUACAACAAAAGCUCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUG ...((........))..........((((.((((...(((((.((((((((((....))))))))))..(((..(((......)))..))).....)))))..)))).)).))....... ( -30.40) >DroSim_CAF1 7189 118 - 1 ACAGGAGAAAAACCCGAAAA--AAAGCGUACAACAAAAGCUCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUG ...((........)).....--...((((.((((...(((((.((((((((((....))))))))))..(((..(((......)))..))).....)))))..)))).)).))....... ( -30.40) >DroEre_CAF1 7320 119 - 1 ACAGGAGAAAAACCCGAA-AAAAAAGCGUACAACAAAAGCUCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUG ...((........))...-......((((.((((...(((((.((((((((((....))))))))))..(((..(((......)))..))).....)))))..)))).)).))....... ( -30.40) >DroYak_CAF1 7199 119 - 1 ACAGGAGAAAAACCCGAA-AAAAAAGCGUACAACAAAAGCUCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUG ...((........))...-......((((.((((...(((((.((((((((((....))))))))))..(((..(((......)))..))).....)))))..)))).)).))....... ( -30.40) >consensus ACAGGAGAAAAACCCGAAAAAAAAAGCGUACAACAAAAGCUCAGCCAAAGCGGAAACCUGUUUUGGCCAUCGAGCAAAGUGAGUUGAGUGAGCAAAGAGUUAAGUUGAGCAGCAGCAUUG ...((........))..........((((.((((...(((((.((((((((((....))))))))))..(((..(((......)))..))).....)))))..)))).)).))....... (-30.40 = -30.40 + 0.00)

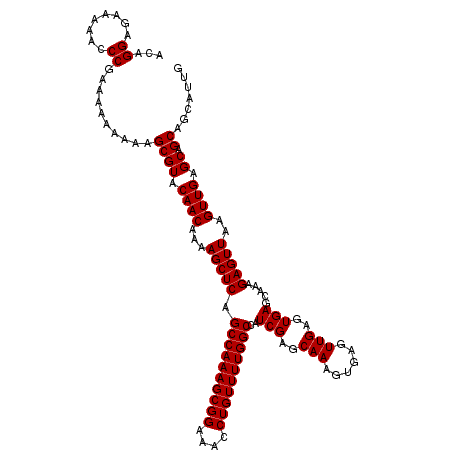
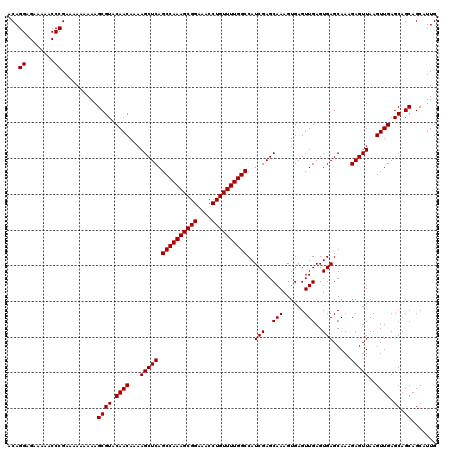
| Location | 9,510,260 – 9,510,379 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -39.48 |
| Energy contribution | -39.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510260 119 - 20766785 GCAGCAGGGAUGUUU-CACUCGAGGGUUGGGAUUCCAGAUAUAUUGGGUAUUGGGCCCAAAGCCCAAGCACACGAAACGAAAAAUAAAACAGCAAACUGUGGGUGAGGCCGCGCUGCUGC (((((((.(..((((-(((((..(((((((....)).......((((((.....))))))))))).......................((((....)))))))))))))..).))))))) ( -45.10) >DroSec_CAF1 7284 119 - 1 GCAGCAGGGAUGUUUCCACUCGAGGGUUGGGAAUCCAGAUAUAUUGGGUAUUGGGCCCA-AGCCCAAGCACACGAAACGAAAAAUAAAACAGCAAACUGUGGGUGAGGCCGCGCUGCUGC (((((((.(..(((((.((((..((((((((...((((.(((.....))))))).))).-))))).......................((((....)))))))))))))..).))))))) ( -41.70) >DroSim_CAF1 7307 119 - 1 GCAGCAGGGAUGUUUCCACUCGAGGGUUGGGAAUCCAGAUAUAUUGGGUAUUGGGCCCA-AGCCCAAGCACACGAAACGAAAAAUAAAACAGCAAACUGUGGGUGAGGCCGCGCUGCUGC (((((((.(..(((((.((((..((((((((...((((.(((.....))))))).))).-))))).......................((((....)))))))))))))..).))))))) ( -41.70) >DroEre_CAF1 7439 118 - 1 GCAGCAGGGAUGUUU-CACUCGAGGGUUGGGAAUCCAGAUAUAUUGGGUAUUGGACCCA-AACCCAAGCACACAAAACGAAAAAUAAAACAACAAACUGUGGGUGAGGCCGCGCUGCUGC (((((((.(..((((-(((((..((((((((..(((((.(((.....))))))))))).-))))).......................(((......))))))))))))..).))))))) ( -43.70) >DroYak_CAF1 7318 118 - 1 GCAUCAGGGAUGUUU-CACUCGAGGGUUGGGAAUCCAGAUAUAUUGGGUAUUGGACCCA-AACCCAAGCACACAAAACGAAAUAUAAAACAGCAAACUGUGGGUGAGGCCGCGCUGCUGC (((.(((.(..((((-(((((..((((((((..(((((.(((.....))))))))))).-))))).......................((((....)))))))))))))..).))).))) ( -39.60) >consensus GCAGCAGGGAUGUUU_CACUCGAGGGUUGGGAAUCCAGAUAUAUUGGGUAUUGGGCCCA_AGCCCAAGCACACGAAACGAAAAAUAAAACAGCAAACUGUGGGUGAGGCCGCGCUGCUGC (((((((.(..((((.(((((..((((((((..(((((.(((.....))))))))))))..)))).......................((((....)))))))))))))..).))))))) (-39.48 = -39.40 + -0.08)

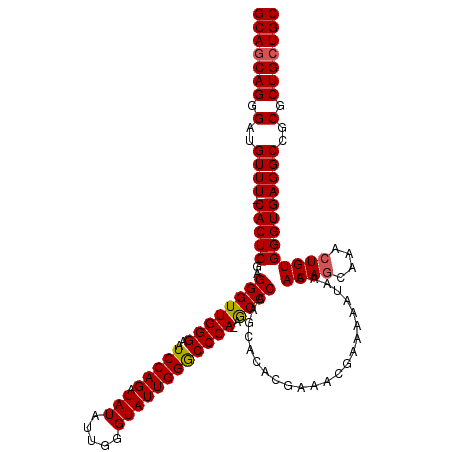
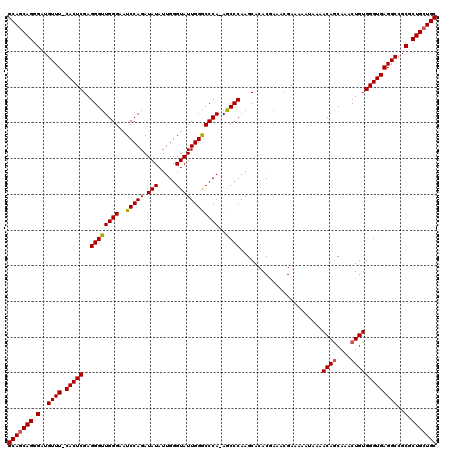
| Location | 9,510,340 – 9,510,459 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -37.96 |
| Energy contribution | -38.64 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510340 119 + 20766785 AUCUGGAAUCCCAACCCUCGAGUG-AAACAUCCCUGCUGCCGCAAAACAGCCCUCAAAAGCGGGCAAUCGCAACCUGAGACACUGGGAAUGCGAGUGCAGAUGGAACUUGGGAAUCGUAU .....((.((((((...((....)-)....(((...((((.((......((((.(....).))))..(((((.(((........)))..)))))))))))..)))..)))))).)).... ( -34.30) >DroSec_CAF1 7363 120 + 1 AUCUGGAUUCCCAACCCUCGAGUGGAAACAUCCCUGCUGCCGCAAAACAGCCCUCAAAAGCGGGCAAUCGCAACCUGAGACACUGGGAAUGCGAGUGCAGAUGGAACUUGGGAAUCGCAU ...(((((((((((........((....))(((...((((.((......((((.(....).))))..(((((.(((........)))..)))))))))))..)))..))))))))).)). ( -42.00) >DroSim_CAF1 7386 120 + 1 AUCUGGAUUCCCAACCCUCGAGUGGAAACAUCCCUGCUGCCGCAAAACAGCCCUCAAAAGCGGGCAAUUGCAACCUGAGACACUGGGAAUGCGAGUGCAGAUGGAACUUGGGAAUCGCAU ...(((((((((((.((..(..((....))..)((((.((((((.....((((.(....).))))........(((........)))..)))).))))))..))...))))))))).)). ( -40.70) >DroEre_CAF1 7518 119 + 1 AUCUGGAUUCCCAACCCUCGAGUG-AAACAUCCCUGCUGCCACAAAACAGCCCUCAAAAGCGGGCAAUCGCAACCUGAGACACUGGGAAUGCGAGUGCAGAUGGAACUUGGGAAUCGCAU ...(((((((((((...((....)-)....(((...((((.((......((((.(....).))))..(((((.(((........)))..)))))))))))..)))..))))))))).)). ( -38.50) >DroYak_CAF1 7397 119 + 1 AUCUGGAUUCCCAACCCUCGAGUG-AAACAUCCCUGAUGCCGCAAAACAGCCCUCAAAAGCGGGCAAUCGCAACCUGAGACACUGGGAAUGCGAGUGCAGAUGGAACUUGGGAAUCGCAU ...(((((((((((((.((.....-...((((...))))..(((.....((((.(....).))))..(((((.(((........)))..))))).))).)).))...))))))))).)). ( -36.60) >consensus AUCUGGAUUCCCAACCCUCGAGUG_AAACAUCCCUGCUGCCGCAAAACAGCCCUCAAAAGCGGGCAAUCGCAACCUGAGACACUGGGAAUGCGAGUGCAGAUGGAACUUGGGAAUCGCAU .....(((((((((.......((....)).(((...((((.((......((((.(....).))))..(((((.(((........)))..)))))))))))..)))..))))))))).... (-37.96 = -38.64 + 0.68)


| Location | 9,510,340 – 9,510,459 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -39.22 |
| Energy contribution | -39.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9510340 119 - 20766785 AUACGAUUCCCAAGUUCCAUCUGCACUCGCAUUCCCAGUGUCUCAGGUUGCGAUUGCCCGCUUUUGAGGGCUGUUUUGCGGCAGCAGGGAUGUUU-CACUCGAGGGUUGGGAUUCCAGAU ....((.((((((.(..(.(((((..(((((....((((.(((((((..(((......))).)))))))))))...)))))..)))))((....)-)....)..).)))))).))..... ( -39.30) >DroSec_CAF1 7363 120 - 1 AUGCGAUUCCCAAGUUCCAUCUGCACUCGCAUUCCCAGUGUCUCAGGUUGCGAUUGCCCGCUUUUGAGGGCUGUUUUGCGGCAGCAGGGAUGUUUCCACUCGAGGGUUGGGAAUCCAGAU .((.(((((((((.(..(..((((..(((((....((((.(((((((..(((......))).)))))))))))...)))))..))))(((....)))....)..).)))))))))))... ( -46.20) >DroSim_CAF1 7386 120 - 1 AUGCGAUUCCCAAGUUCCAUCUGCACUCGCAUUCCCAGUGUCUCAGGUUGCAAUUGCCCGCUUUUGAGGGCUGUUUUGCGGCAGCAGGGAUGUUUCCACUCGAGGGUUGGGAAUCCAGAU .((.(((((((((.(..(..((((....((((.....)))).....(((((((..((((........))))....))))))).))))(((....)))....)..).)))))))))))... ( -44.30) >DroEre_CAF1 7518 119 - 1 AUGCGAUUCCCAAGUUCCAUCUGCACUCGCAUUCCCAGUGUCUCAGGUUGCGAUUGCCCGCUUUUGAGGGCUGUUUUGUGGCAGCAGGGAUGUUU-CACUCGAGGGUUGGGAAUCCAGAU .((.(((((((((.(..(...(((....))).((((....(((((((..(((......))).)))))))((((((....)))))).)))).....-.....)..).)))))))))))... ( -42.70) >DroYak_CAF1 7397 119 - 1 AUGCGAUUCCCAAGUUCCAUCUGCACUCGCAUUCCCAGUGUCUCAGGUUGCGAUUGCCCGCUUUUGAGGGCUGUUUUGCGGCAUCAGGGAUGUUU-CACUCGAGGGUUGGGAAUCCAGAU .((.(((((((((........(((....)))..(((((..((((..(((((((..((((........))))....)))))))....))))..))(-(....))))))))))))))))... ( -41.00) >consensus AUGCGAUUCCCAAGUUCCAUCUGCACUCGCAUUCCCAGUGUCUCAGGUUGCGAUUGCCCGCUUUUGAGGGCUGUUUUGCGGCAGCAGGGAUGUUU_CACUCGAGGGUUGGGAAUCCAGAU ....(((((((((.(..(..((((..(((((....((((.(((((((..(((......))).)))))))))))...)))))..))))(..((....))..))..).)))))))))..... (-39.22 = -39.66 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:53 2006