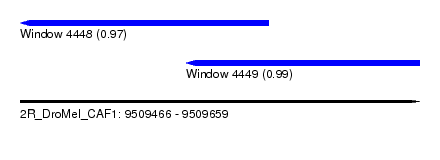
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,509,466 – 9,509,659 |
| Length | 193 |
| Max. P | 0.988450 |

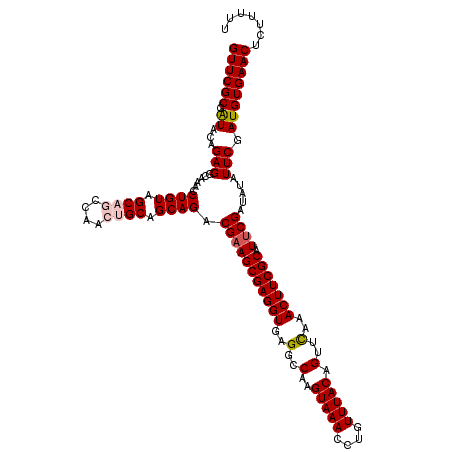
| Location | 9,509,466 – 9,509,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -31.60 |
| Energy contribution | -32.28 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9509466 120 - 20766785 GUUCGCGAUACGGAGGCAAACCUGUAGCAGCCAACUGCAGCAGACGAAGCGAGGUGAGGCCAAGUAAACCUGUUUACAGUUCAAACUUCGCAUUCGAUAUAUUCGAUGUGAACUCUUUUU (((((((...((((.......((((.((((....)))).)))).(((((((((((..(..(..(((((....))))).)..)..))))))).)))).....)))).)))))))....... ( -37.90) >DroSec_CAF1 6474 120 - 1 GUUCGCGAUACAGAGGCAAACCUGUAGCAGCCAACUGCAGCAGACGAAGCGAGGUGAGGCCAAGUAAACCUGUUUACAGUUUAAACUUCGCAUUCGAUAUAUUCGAUGUGAACUCUUUUU (((((((..............((((.((((....)))).)))).(((((((((((.((((...(((((....))))).))))..))))))).))))..........)))))))....... ( -35.70) >DroSim_CAF1 6483 120 - 1 GUUCGCGAUACAGAGGCAAACCUGUAGCAGCCAACUGCAGCAGACGUAGCGAGGUGAGGCCAAGUAAACCUGUUUACAGUUCAAACUUCGCAUUCGAUAUAUUCGAUGUGAACUCUUUUU (((((((..............((((.((((....)))).)))).....(((((((..(..(..(((((....))))).)..)..)))))))..((((.....)))))))))))....... ( -35.00) >DroEre_CAF1 6486 120 - 1 GUUCGCGGUAAAGAGGAAAACCUGUAGCUACCAACUGCAGCAGACGAAGCGAGGUGAGGCCAAGUAAACCUGUUUACAGUUCAAACUUCGCAUUCGAUAUAUUCGAUGUGAACUCUUUUU (((((((.....(((......((((.((........)).)))).(((((((((((..(..(..(((((....))))).)..)..))))))).)))).....)))..)))))))....... ( -33.60) >DroYak_CAF1 6394 120 - 1 GUUCGCUAUACAGAGGAGUACCUGUAGCUACCAACUGCAGCAGACGAAGCGAGGUGAGGCCAAGUAAACCUGUUUACAGUUCAAACUUCGCAUUCGAUAUAUUCGAUGUGAACUCUUUUU ((((((.((...(((......((((.((........)).)))).(((((((((((..(..(..(((((....))))).)..)..))))))).)))).....))).))))))))....... ( -32.60) >consensus GUUCGCGAUACAGAGGCAAACCUGUAGCAGCCAACUGCAGCAGACGAAGCGAGGUGAGGCCAAGUAAACCUGUUUACAGUUCAAACUUCGCAUUCGAUAUAUUCGAUGUGAACUCUUUUU ((((((.((...(((......((((.((((....)))).)))).(((((((((((..(..(..(((((....))))).)..)..))))))).)))).....))).))))))))....... (-31.60 = -32.28 + 0.68)

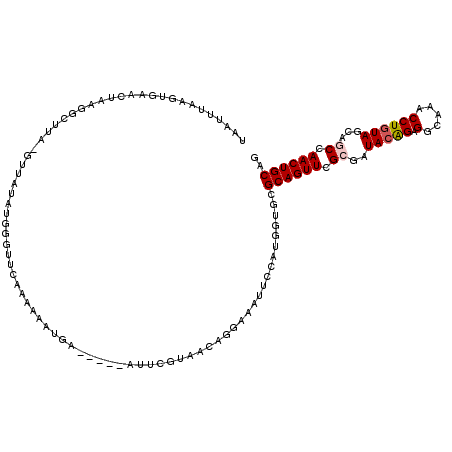
| Location | 9,509,546 – 9,509,659 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9509546 113 - 20766785 UAACUUAAGUGAAAUAAGGCUUAUGUUAUAUGGAUUCAAAA-AUGA-----AUGAAU-ACAGGAAAUUCCAUGGCGCGCAGUUCGCGAUACGGAGGCAAACCUGUAGCAGCCAACUGCAG ((((.(((((........))))).)))).....(((((...-.)))-----))...(-(((((...((((.((.((((.....)))).)).)))).....))))))((((....)))).. ( -28.60) >DroSec_CAF1 6554 115 - 1 UAAUUUAAGUGGAUAAAGGCUUAAGUUAUAUGGGUUCAAAAAAUGA-----AUUCGUAACAGGAAAUUCCAUGGUGCGCAGUUCGCGAUACAGAGGCAAACCUGUAGCAGCCAACUGCAG ((((((((((........))))))))))((((((((((.....)))-----))))))).((((.....)).))....((((((.((..(((((.(.....))))))...)).)))))).. ( -35.10) >DroSim_CAF1 6563 115 - 1 UAAUUUAAGUGAACAAAGGCUUAAGUUAUAUGGGUUCAAAAAAUGA-----AUUCGUAACAGGAAAUUCCAUGGUGCGCAGUUCGCGAUACAGAGGCAAACCUGUAGCAGCCAACUGCAG ((((((((((........))))))))))((((((((((.....)))-----))))))).((((.....)).))....((((((.((..(((((.(.....))))))...)).)))))).. ( -35.10) >DroEre_CAF1 6566 105 - 1 UGUUUA-CGUAAACUACG--------------GGUUCAACAAAUGAAACAAAUUCGUUACAGAAAAUUCCACGGGCAGCAGUUCGCGGUAAAGAGGAAAACCUGUAGCUACCAACUGCAG (((...-.(((..(((((--------------((((...(.((((((.....))))))..........((.(((((....))))).))......)...))))))))).))).....))). ( -23.00) >DroYak_CAF1 6474 110 - 1 UGAUUU-AGUGAACUAGGACU--------AU-AGUUCCAAAAAUUAAACAUAUUUGUUACAGAAACUUCAAUGGUCCGCAGUUCGCUAUACAGAGGAGUACCUGUAGCUACCAACUGCAG .....(-((((((((.(((((--------((-.((..((((.((.....)).))))..)).((....)).)))))))..)))))))))(((((.(.....))))))((........)).. ( -29.00) >consensus UAAUUUAAGUGAACUAAGGCUUA_GUUAUAUGGGUUCAAAAAAUGA_____AUUCGUAACAGGAAAUUCCAUGGUGCGCAGUUCGCGAUACAGAGGCAAACCUGUAGCAGCCAACUGCAG .............................................................................((((((.((..(((((.(.....))))))...)).)))))).. (-14.50 = -14.94 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:48 2006