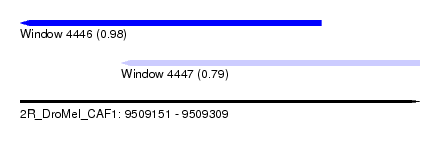
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,509,151 – 9,509,309 |
| Length | 158 |
| Max. P | 0.984480 |

| Location | 9,509,151 – 9,509,270 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.16 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
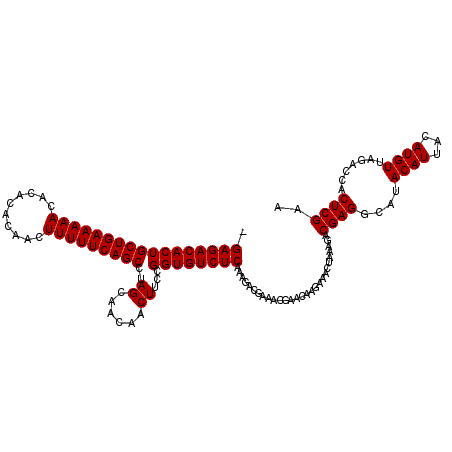
>2R_DroMel_CAF1 9509151 119 - 20766785 -GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAAACUAAAGACGAGGCAUACAUUACAUGUUAGACCACUCGAA -(((((((((((((((((..........)))))))))..((......))...))))))))............................((((....((((...)))).......)))).. ( -23.50) >DroSec_CAF1 6159 119 - 1 -GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAAACUAAAGACGAGGCAUACAUUACAUGUUAGACCACUCGAA -(((((((((((((((((..........)))))))))..((......))...))))))))............................((((....((((...)))).......)))).. ( -23.50) >DroSim_CAF1 6168 119 - 1 -GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAAACUAAAGACGAGGCAUACAUUACAUGUUAGACCACUCGAA -(((((((((((((((((..........)))))))))..((......))...))))))))............................((((....((((...)))).......)))).. ( -23.50) >DroEre_CAF1 6169 120 - 1 GGAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAACCGAACAAGAAACUAAAGACGAGGCAUACAUUACAUGUUAGACCACUCGAA .(((((((((((((((((..........)))))))))..((......))...))))))))............................((((....((((...)))).......)))).. ( -23.50) >DroYak_CAF1 6076 119 - 1 -GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAACCGAACAAGAAACUAAAGACGAGGCAUACAUUACAUGUUAGACCACUCGAA -(((((((((((((((((..........)))))))))..((......))...))))))))............................((((....((((...)))).......)))).. ( -23.50) >consensus _GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAAACUAAAGACGAGGCAUACAUUACAUGUUAGACCACUCGAA .(((((((((((((((((..........)))))))))..((......))...))))))))............................((((....((((...)))).......)))).. (-23.50 = -23.50 + 0.00)

| Location | 9,509,191 – 9,509,309 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
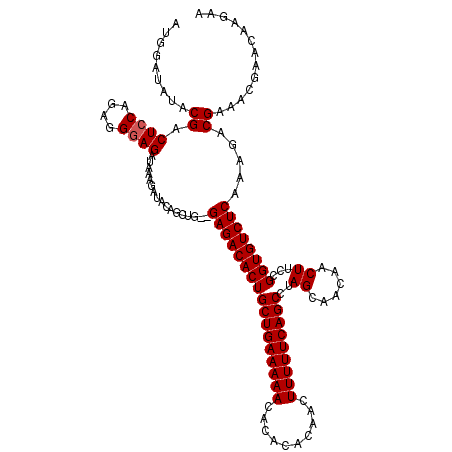
>2R_DroMel_CAF1 9509191 118 - 20766785 AUGGAUAUACGGCUCCAGAGGGAGAUAAAGAUACAGCUG--GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAA .(((((((.(((((((....))))...........((((--(.......(((((((((..........))))))))))))))........)))))))).))................... ( -25.81) >DroSec_CAF1 6199 118 - 1 GUGGAUAUACGACUCCAGAGGGAGAUAAAGAUACAGAUG--GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAA .........((.((((....))))...............--(((((((((((((((((..........)))))))))..((......))...)))))))).....))............. ( -25.00) >DroSim_CAF1 6208 118 - 1 AUGGAUAUACGACUCCAGAGGGAGAUAAAGAUACAGCUG--GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAA .........((.((((....))))...............--(((((((((((((((((..........)))))))))..((......))...)))))))).....))............. ( -25.00) >DroEre_CAF1 6209 120 - 1 AUGGAUAUACGGCUGCAGAGGGAGAUAAAGAUACAGCUGGGGAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAACCGAACAAGAA .(((.....((((((..................))))))..(((((((((((((((((..........)))))))))..((......))...)))))))).........)))........ ( -27.97) >DroYak_CAF1 6116 118 - 1 AUGGAUAUACGUCUCCAGAGGGAGAUAAAGAUACAGCUG--GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAACCGAACAAGAA ..(((.....((((((((..(............)..)))--)))))...(((((((((..........)))))))))............)))((((((.....)))..)))......... ( -31.10) >consensus AUGGAUAUACGACUCCAGAGGGAGAUAAAGAUACAGCUG__GAGACACUGCUGAAAAACACACACAACUUUUUCAGCCUAGCAACAACUUCCGGUGUCUCAAAGACGAAACGAACAAGAA .........((.((((....)))).................(((((((((((((((((..........)))))))))..((......))...)))))))).....))............. (-23.44 = -23.64 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:46 2006