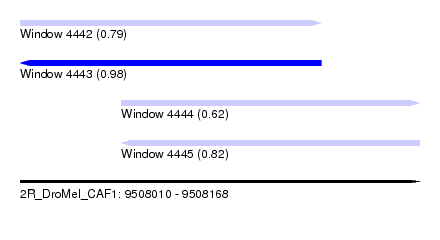
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,508,010 – 9,508,168 |
| Length | 158 |
| Max. P | 0.981715 |

| Location | 9,508,010 – 9,508,129 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9508010 119 + 20766785 GGUAAGUAAGUUAAGUCUCUUAUCGCCUAAUUAGUUUAUAGCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUCAUAUGCCUUUU-UUUCUCU ((((.(((((..(((((.((((.(((..((((((...........))))))..))))))).)))))))))).....((((((.(........).))))))....))))....-....... ( -21.80) >DroSec_CAF1 5023 120 + 1 GGUAAGUAAGUUAACGCUCUUAUCGCCUAAUUAGUUUAUAGCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUACUUCGGGUUUUGAUAUGCCUUUUUUUUCACU ((((.(((((....(((((....(((..((((((...........))))))..))).)))))....)))))...((((((((.(........).))))))))..))))............ ( -25.60) >DroSim_CAF1 5022 119 + 1 GGUAAGUAAGUUAACGCUCUUAUCGCCUAAUUAGUUUAUAGCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAUAUGCCUUUU-UUUCUCU ((((.(((((....(((((....(((..((((((...........))))))..))).)))))....)))))...((((((((.(........).))))))))..))))....-....... ( -25.60) >DroEre_CAF1 5047 116 + 1 GGUAAGUAAGCUAAUGCUCUUAUCGCCUAAUUAGUUAAUAGCGUUUUAUUUUUGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAAAUGCCUUU--UUU--CU ((((((..(((....)))))))))(((((....(..(((((....)))))..)...)))))........(((((((((((((.(........).)))))))))))))....--...--.. ( -30.10) >DroYak_CAF1 4986 117 + 1 GGUAAGUAAGUUAACGCUCUUAUCGCCUAAUUAGUUUAUAGCGUUUUAUUUUCUUGUGGGUGGCUUUUCGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAAAUGCCUUUU-UUU--CU ((((((...(....)...))))))(((((...((...((((....))))...))..)))))........(((((((((((((.(........).))))))))))))).....-...--.. ( -28.60) >consensus GGUAAGUAAGUUAACGCUCUUAUCGCCUAAUUAGUUUAUAGCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAUAUGCCUUUU_UUUC_CU ((((((..(((....)))))))))(((((...........(((.........))).)))))........((((.((((((((.(........).)))))))).))))............. (-20.84 = -21.16 + 0.32)

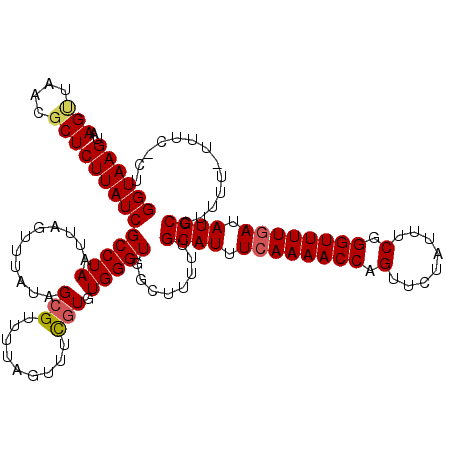
| Location | 9,508,010 – 9,508,129 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9508010 119 - 20766785 AGAGAAA-AAAAGGCAUAUGAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGCUAUAAACUAAUUAGGCGAUAAGAGACUUAACUUACUUACC .(((...-.....((((.(.((((((.(........).)))))).).))))..........................((((............))))........)))............ ( -14.50) >DroSec_CAF1 5023 120 - 1 AGUGAAAAAAAAGGCAUAUCAAAACCCGAAGUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGCUAUAAACUAAUUAGGCGAUAAGAGCGUUAACUUACUUACC (((((........((((.((((((((.(........).)))))))).))))........................((((((.....((....))........))))))...))))).... ( -22.12) >DroSim_CAF1 5022 119 - 1 AGAGAAA-AAAAGGCAUAUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGCUAUAAACUAAUUAGGCGAUAAGAGCGUUAACUUACUUACC .(((...-.....((((.((((((((.(........).)))))))).))))........................((((((.....((....))........))))))..)))....... ( -21.02) >DroEre_CAF1 5047 116 - 1 AG--AAA--AAAGGCAUUUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACAAAAAUAAAACGCUAUUAACUAAUUAGGCGAUAAGAGCAUUAGCUUACUUACC ..--...--.((((((((((((((((.(........).)))))))))))))..........................((((............))))....((((....)))).)))... ( -24.70) >DroYak_CAF1 4986 117 - 1 AG--AAA-AAAAGGCAUUUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCGAAAAGCCACCCACAAGAAAAUAAAACGCUAUAAACUAAUUAGGCGAUAAGAGCGUUAACUUACUUACC ..--...-..((((((((((((((((.(........).)))))))))))))........................((((((.....((....))........))))))..)))....... ( -24.32) >consensus AG_GAAA_AAAAGGCAUAUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGCUAUAAACUAAUUAGGCGAUAAGAGCGUUAACUUACUUACC ..........(((((((.((((((((.(........).)))))))).))))..........................((((............)))).................)))... (-17.30 = -17.50 + 0.20)

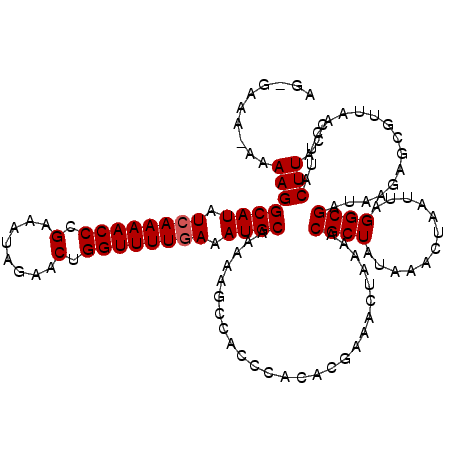
| Location | 9,508,050 – 9,508,168 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9508050 118 + 20766785 GCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUCAUAUGCCUUUU-UUUCUCUUCUC-AUGUUGAUAAGACCUUGAAGCGCUUUUCGUGUAGA (((((((((...((((((((..((.....))...))((((((.(........).))))))))))))......-....((((.((-.....)).))))..)))))))))............ ( -21.90) >DroSec_CAF1 5063 119 + 1 GCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUACUUCGGGUUUUGAUAUGCCUUUUUUUUCACUUCUC-AUGUUGAUAGGACCUUGAAGCGCUUUUCGUGUAGA ((((((((((((((((.((((((......((((.((((((((.(........).)))))))).))))........))))))..)-)))......))))..))))))))............ ( -29.04) >DroSim_CAF1 5062 119 + 1 GCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAUAUGCCUUUU-UUUCUCUUCUCUAUGUUGAUAAGACCUUGAAGCGCUUUUCGUGUAGA (((.....((((((...((..........((((.((((((((.(........).)))))))).)))).....-....((((.((......)).)))))).))))))......)))..... ( -27.00) >DroEre_CAF1 5087 115 + 1 GCGUUUUAUUUUUGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAAAUGCCUUU--UUU--CUUCUG-AUGUUGAUAAGACCUUGAAGCGCUUUUCGUGUAGA ..........((((..((((.(((.....(((((((((((((.(........).)))))))))))))....--...--((((..-..(((.....)))...)))).))).))))..)))) ( -28.80) >DroYak_CAF1 5026 116 + 1 GCGUUUUAUUUUCUUGUGGGUGGCUUUUCGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAAAUGCCUUUU-UUU--CUUGUG-AUGUUGAUAAGACCUUGAAGUGCUUUUCGUGUAGA ...........(((..((((.(((.(((((((((((((((((.(........).))))))))))))).....-..(--(((((.-......))))))....)))).))).))))...))) ( -28.80) >consensus GCGUUUUAGUUUCGUGUGGGUGGCUUUUUGCAUUUCAAAACCAGUUCUAUUUCGGGUUUUGAUAUGCCUUUU_UUUC_CUUCUC_AUGUUGAUAAGACCUUGAAGCGCUUUUCGUGUAGA ((((((((.........((((........((((.((((((((.(........).)))))))).))))...........(((.((......)).)))))))))))))))............ (-21.96 = -22.24 + 0.28)


| Location | 9,508,050 – 9,508,168 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9508050 118 - 20766785 UCUACACGAAAAGCGCUUCAAGGUCUUAUCAACAU-GAGAAGAGAAA-AAAAGGCAUAUGAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGC ............(((.(((....((((((....))-))))...))).-.....((((.(.((((((.(........).)))))).).))))..........................))) ( -16.40) >DroSec_CAF1 5063 119 - 1 UCUACACGAAAAGCGCUUCAAGGUCCUAUCAACAU-GAGAAGUGAAAAAAAAGGCAUAUCAAAACCCGAAGUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGC ......((.....((((((...((.......))..-..)))))).........((((.((((((((.(........).)))))))).))))...............))............ ( -20.90) >DroSim_CAF1 5062 119 - 1 UCUACACGAAAAGCGCUUCAAGGUCUUAUCAACAUAGAGAAGAGAAA-AAAAGGCAUAUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGC ............(.((((.....((((.((........)).))))..-.....((((.((((((((.(........).)))))))).))))...)))))..................... ( -20.40) >DroEre_CAF1 5087 115 - 1 UCUACACGAAAAGCGCUUCAAGGUCUUAUCAACAU-CAGAAG--AAA--AAAGGCAUUUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACAAAAAUAAAACGC ............(((((((...((.......))..-..))))--...--....(((((((((((((.(........).)))))))))))))..........................))) ( -21.80) >DroYak_CAF1 5026 116 - 1 UCUACACGAAAAGCACUUCAAGGUCUUAUCAACAU-CACAAG--AAA-AAAAGGCAUUUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCGAAAAGCCACCCACAAGAAAAUAAAACGC (((....(((......)))..((.(((.((....(-(....)--)..-.....(((((((((((((.(........).))))))))))))))).))))).......)))........... ( -23.80) >consensus UCUACACGAAAAGCGCUUCAAGGUCUUAUCAACAU_GAGAAG_GAAA_AAAAGGCAUAUCAAAACCCGAAAUAGAACUGGUUUUGAAAUGCAAAAAGCCACCCACACGAAACUAAAACGC .......((.(((.(((....)))))).)).......................((((.((((((((.(........).)))))))).))))............................. (-15.78 = -16.02 + 0.24)

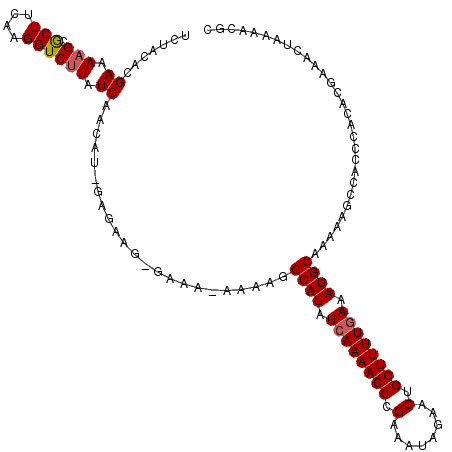

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:44 2006