| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,507,850 – 9,507,970 |
| Length | 120 |
| Max. P | 0.938495 |

| Location | 9,507,850 – 9,507,970 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
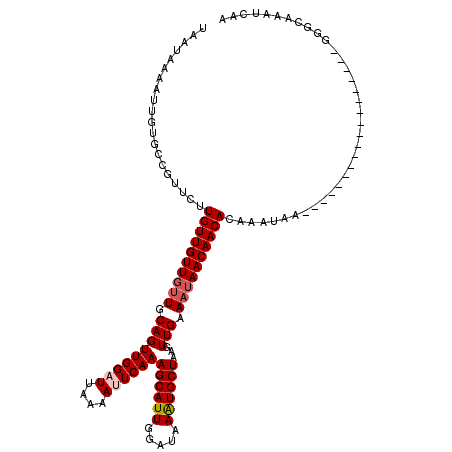
>2R_DroMel_CAF1 9507850 120 + 20766785 UAAUAAAAUUGUGCCGUUCUUGUUGUUGUUGCAGUUGGAUUAAAAUUCAAAGCAUUGGAUAAAUGCUAAGUUGAAAUAACAACAGUAAUAACAAAUGGAAAAUAGCAAAUAGCAAAUCAA ........((((....((((..((((((((((..((((((....))))))((((((.....))))))..((((......)))).))))))))))..))))....))))............ ( -28.10) >DroSec_CAF1 4881 102 + 1 UAAUAAAGUUGUGCCGUUCUUGUUGUUGUUGCAGUUGGAUUAAAAGUCAAAGCAUUGGAUAAAUGCUAAGUUGAAAUAACAACACAAAUAA------------------GGGCAAAUCAA .......(...((((.((..((((((((((.(((...(((.....)))..((((((.....))))))...))).)))))))))).....))------------------.))))...).. ( -24.00) >DroSim_CAF1 4880 102 + 1 UAAUAAAAUUGUGCCGUUCUUGUUGUUGUUGCAGUUGGAUUAAAAGUCAAAGCAUUGGAUAAAUGCUAAGUUGAAAUAACAACACAAAUAA------------------GGGCAAAUCAA ...........((((.((..((((((((((.(((...(((.....)))..((((((.....))))))...))).)))))))))).....))------------------.))))...... ( -23.50) >DroEre_CAF1 4906 101 + 1 UAAUAAAAUUGUGCCGUUCUUGUUGUUGUUGCAGUUGGAUUAAAAUUCAAAGCAUUU-AUCAGUGCUAAAUUGAAUGAACAACAAAAGUAA------------------GGGCAAAUCAU ...........((((.(((((.((((((((.((.((((((....))))))(((((..-....)))))........))))))))))))).))------------------.))))...... ( -24.70) >DroYak_CAF1 4845 101 + 1 UAAUAAAAUUGUGUCGUUCUUGUUGUUGUUGCAGUUGGAUUAAAAUUCAAAGCAUUU-AUCAGUGCUAAAUUGAAAUAACAACACAAGUAA------------------GGGUAAAUCAU ...........(..(.(((((((.(((((((...((((((....))))))(((((..-....))))).........)))))))))))).))------------------.)..)...... ( -20.60) >consensus UAAUAAAAUUGUGCCGUUCUUGUUGUUGUUGCAGUUGGAUUAAAAUUCAAAGCAUUGGAUAAAUGCUAAGUUGAAAUAACAACACAAAUAA__________________GGGCAAAUCAA ....................((((((((((.(((((((((....))))))((((((.....))))))...))).)))))))))).................................... (-16.66 = -17.22 + 0.56)

| Location | 9,507,850 – 9,507,970 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -16.48 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
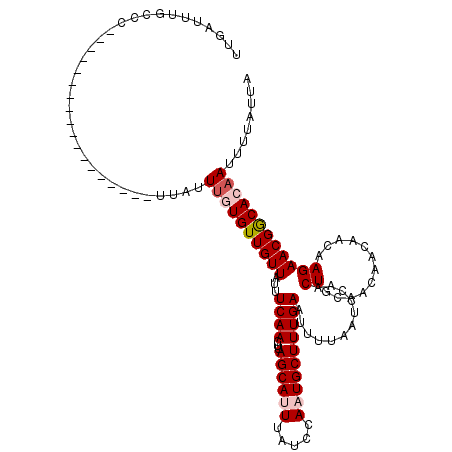
>2R_DroMel_CAF1 9507850 120 - 20766785 UUGAUUUGCUAUUUGCUAUUUUCCAUUUGUUAUUACUGUUGUUAUUUCAACUUAGCAUUUAUCCAAUGCUUUGAAUUUUAAUCCAACUGCAACAACAACAAGAACGGCACAAUUUUAUUA .............(((..((((....(((((.((.(.(((((((.(((((...((((((.....)))))))))))...)))..)))).).)).)))))..))))..)))........... ( -16.40) >DroSec_CAF1 4881 102 - 1 UUGAUUUGCCC------------------UUAUUUGUGUUGUUAUUUCAACUUAGCAUUUAUCCAAUGCUUUGACUUUUAAUCCAACUGCAACAACAACAAGAACGGCACAACUUUAUUA ......((((.------------------((.(((((((((((...((((...((((((.....)))))))))).........(....).)))))).))))))).))))........... ( -17.00) >DroSim_CAF1 4880 102 - 1 UUGAUUUGCCC------------------UUAUUUGUGUUGUUAUUUCAACUUAGCAUUUAUCCAAUGCUUUGACUUUUAAUCCAACUGCAACAACAACAAGAACGGCACAAUUUUAUUA ......((((.------------------((.(((((((((((...((((...((((((.....)))))))))).........(....).)))))).))))))).))))........... ( -17.00) >DroEre_CAF1 4906 101 - 1 AUGAUUUGCCC------------------UUACUUUUGUUGUUCAUUCAAUUUAGCACUGAU-AAAUGCUUUGAAUUUUAAUCCAACUGCAACAACAACAAGAACGGCACAAUUUUAUUA ......((((.------------------((.((((((((((((((((((...((((.....-...)))))))))............)).)))))))).))))).))))........... ( -17.60) >DroYak_CAF1 4845 101 - 1 AUGAUUUACCC------------------UUACUUGUGUUGUUAUUUCAAUUUAGCACUGAU-AAAUGCUUUGAAUUUUAAUCCAACUGCAACAACAACAAGAACGACACAAUUUUAUUA ...........------------------....((((((((((..(((((...((((.....-...)))))))))...........((............))))))))))))........ ( -14.40) >consensus UUGAUUUGCCC__________________UUAUUUGUGUUGUUAUUUCAACUUAGCAUUUAUCCAAUGCUUUGAAUUUUAAUCCAACUGCAACAACAACAAGAACGGCACAAUUUUAUUA .................................((((((((((...((((...((((((.....))))))))))............((............))))))))))))........ (-12.08 = -12.92 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:40 2006