
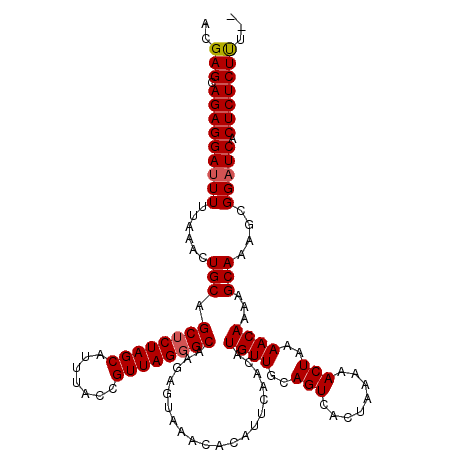
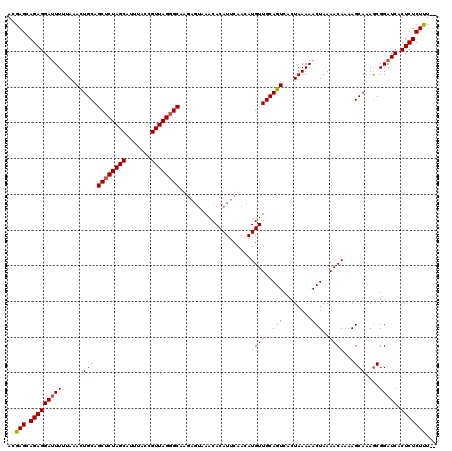
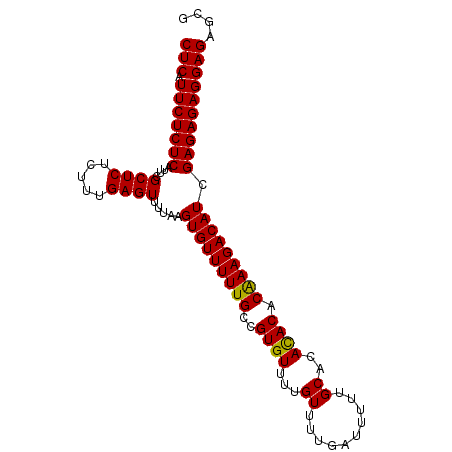
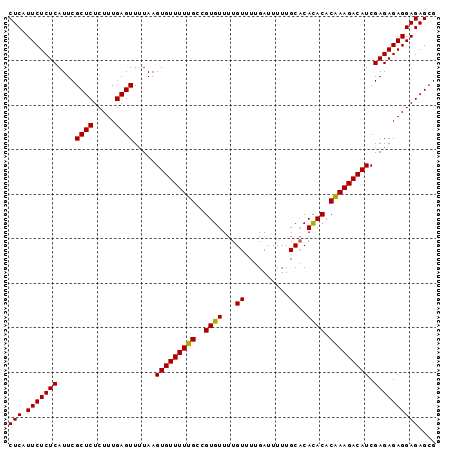
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,507,596 – 9,507,810 |
| Length | 214 |
| Max. P | 0.971231 |

| Location | 9,507,596 – 9,507,714 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.54 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9507596 118 + 20766785 ACGAGCAGAGGAUUUUUAAACUGCAGCUCUAGCAUUUACCGUUAGGGCAAGAGUAAACACAUUCAACAUGUUGCAGUCACUAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUU-- ..(((.(((((((((....((((((((((((((.......))))))))..((((......)))).......))))))....................((...))))))).))))))).-- ( -28.10) >DroSec_CAF1 4627 118 + 1 ACGAGCAGAGGAUUUUUAAACUGCAGCUCUAGCAUUUACCGUUAGGGCAAGAGUAAACACAUUCAACAUGUUGCAGUCACUAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUU-- ..(((.(((((((((....((((((((((((((.......))))))))..((((......)))).......))))))....................((...))))))).))))))).-- ( -28.10) >DroSim_CAF1 4626 118 + 1 ACGAGCAGAGGAUUUUUAAACUGCAGCUCUAGCAUUUACCGUUAGGGCAAGAGUAAACACAUUCAACAUGUUGCAGUCACUAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUU-- ..(((.(((((((((....((((((((((((((.......))))))))..((((......)))).......))))))....................((...))))))).))))))).-- ( -28.10) >DroYak_CAF1 4605 114 + 1 ACGAGCAGAGGACUUUUAAAUUGCAGCACUAGCAUUUACCGUUAGGGCA------AACACAUUCAACAUGUUGCAGUCACUAAAAACUAAAACAAAAGCAAAACGGAUCACUCUCUCUUG ..(((.((((((((.((...((((.((.(((((.......))))).)).------.............((((..(((........)))..))))...)))))).)).)).)))))))... ( -22.30) >consensus ACGAGCAGAGGAUUUUUAAACUGCAGCUCUAGCAUUUACCGUUAGGGCAAGAGUAAACACAUUCAACAUGUUGCAGUCACUAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUU__ ..(((.(((((((((......(((.((((((((.......))))))))....................((((..(((........)))..))))...)))....))))).)))))))... (-24.06 = -24.38 + 0.31)

| Location | 9,507,676 – 9,507,772 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 99.31 |
| Mean single sequence MFE | -19.31 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.31 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9507676 96 + 20766785 UAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUUCUCAUUCUCUCAUUCGCUCUCUUUGAGUUAUAAGUGUUUUUGCCGUGUUUUGUUUUGA .......((((((((((.((..(((((.((((......((((...................)))).....))))..)))))..)))))))))))). ( -19.31) >DroSec_CAF1 4707 96 + 1 UAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUUCUCAUUCUCUCAUUCGCUCUCUUUGAGUUUUAAGUGUUUUUGCCGUGUUUUGUUUUGA .......((((((((((.((..(((((.((((......((((...................)))).....))))..)))))..)))))))))))). ( -19.31) >DroSim_CAF1 4706 96 + 1 UAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUUCUCAUUCUCUCAUUCGCUCUCUUUGAGUUUUAAGUGUUUUUGCCGUGUUUUGUUUUGA .......((((((((((.((..(((((.((((......((((...................)))).....))))..)))))..)))))))))))). ( -19.31) >consensus UAAAAACUAAAACAAAAGCAAAGCGGAUCACUCUCUUUCUCAUUCUCUCAUUCGCUCUCUUUGAGUUUUAAGUGUUUUUGCCGUGUUUUGUUUUGA .......((((((((((.((..(((((.((((......((((...................)))).....))))..)))))..)))))))))))). (-19.31 = -19.31 + 0.00)

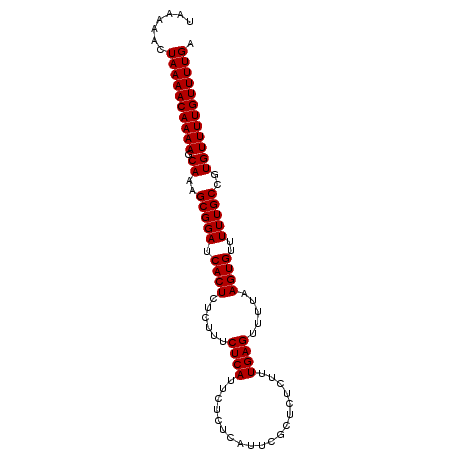
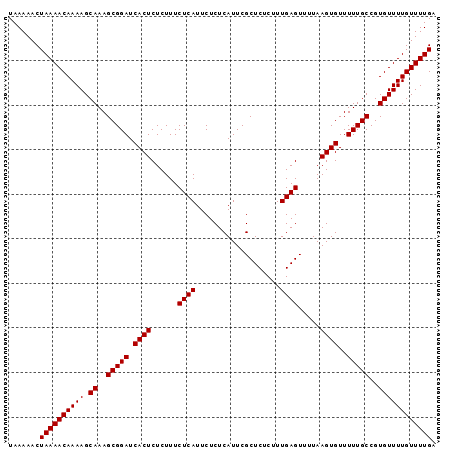
| Location | 9,507,714 – 9,507,810 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9507714 96 + 20766785 CUCAUUCUCUCAUUCGCUCUCUUUGAGUUAUAAGUGUUUUUGCCGUGUUUUGUUUUGAUUUUUGCACAUACACGAAGACAUCGAGAGAGGAGAGCG (((.(((((((....((((.....)))).....(((((((...(((((..(((..........)))...)))))))))))).)))))))))).... ( -26.10) >DroSec_CAF1 4745 96 + 1 CUCAUUCUCUCAUUCGCUCUCUUUGAGUUUUAAGUGUUUUUGCCGUGUUUUGUUUUGAUUUUUGCACACACACAAAGACAUCGAGAGAGGAGAGCG (((.(((((((....((((.....)))).....(((((((((..((((..(((..........))).)))).))))))))).)))))))))).... ( -28.00) >DroSim_CAF1 4744 96 + 1 CUCAUUCUCUCAUUCGCUCUCUUUGAGUUUUAAGUGUUUUUGCCGUGUUUUGUUUUGAUUUUUGCCCACACACAAAGACAUCGAGAGAGGAGAGCG (((.(((((((....((((.....)))).....(((((((((..((((...((..........))..)))).))))))))).)))))))))).... ( -26.60) >consensus CUCAUUCUCUCAUUCGCUCUCUUUGAGUUUUAAGUGUUUUUGCCGUGUUUUGUUUUGAUUUUUGCACACACACAAAGACAUCGAGAGAGGAGAGCG (((.(((((((....((((.....)))).....(((((((((..((((...((..........))..)))).))))))))).)))))))))).... (-26.31 = -25.87 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:38 2006