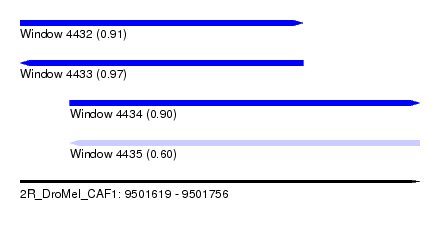
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,501,619 – 9,501,756 |
| Length | 137 |
| Max. P | 0.969589 |

| Location | 9,501,619 – 9,501,716 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -32.59 |
| Energy contribution | -32.15 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
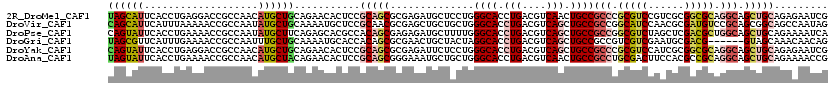
>2R_DroMel_CAF1 9501619 97 + 20766785 --CU------------------GUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGACGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAGCAUCUCGCGCUGCGG --((------------------(..(((.((((((((((........((((((((.((.(((((.....)))))))))))))))........))))...))))))....)))..))) ( -44.29) >DroVir_CAF1 2249 93 + 1 UGCU------------------GAU------GUGUGGCUAUUGGCUGCCGCUGCGGACAUCGCGUUGGAUGCCGGCGGCAGCUGACGUCAGGUGCCCAGCAGCAGCUCGCGUUGCGG ((((------------------(..------.(.((((..(..((((((((..(((.((((......)))))))))))))))..).)))).)....)))))(((((....))))).. ( -44.00) >DroPse_CAF1 2065 88 + 1 --UU------------------GUGGUA---------UGAUUUUCUGCAGCUGCCAGCGUCGAGCUAGACGCCGGCGGCAGCUGACGUCAGGUGCCCAAAAGCAUCUCUCGCUGUGG --..------------------......---------.......(..((((.(((.(((((......))))).)))((((.(((....))).))))..............))))..) ( -35.80) >DroEre_CAF1 2398 97 + 1 --CU------------------GUGGUGAGAGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCAAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAAGAGCAUCUCGCGCUGCGG --((------------------(..(((.(((.((((((....((.((.((((((((((((.....)).)))))))))).)).))))))))((((......))))))).)))..))) ( -44.50) >DroYak_CAF1 2030 97 + 1 --CU------------------GUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGCUGACGUCAGGUGCCCAGGAGAAUCUCGCGCUGCGG --((------------------(..(((.((.....))((((((((.((((((((.((.(((((.....)))))))))))))))......((...)).))))))))...)))..))) ( -46.20) >DroAna_CAF1 2098 115 + 1 --CUGUGUGUGUGACUGAUCCUGUGGUGUGUGUGUGGCGGUUUUCUGCAGCUGCCUGCGGCGUGGAAGUCGCAGGCGGCAGUUGACGUCAGGUGCCCAGCAGCAUUUCCCGCUGCGG --(((((((.(((.(((...(((.((..(.(((((.((((....)))).((((((((((((......)))))))))))).....))).)).)..)))))))))))....))).)))) ( -50.40) >consensus __CU__________________GUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGCUGACGUCAGGUGCCCAGGAGCAUCUCGCGCUGCGG ............................................(((((((.(((((((.(......).)))))))((((.(((....))).))))..............))))))) (-32.59 = -32.15 + -0.44)

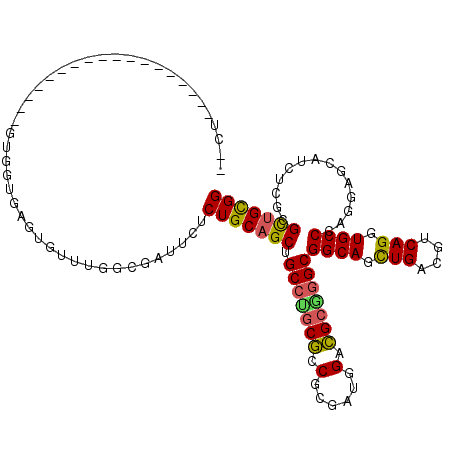
| Location | 9,501,619 – 9,501,716 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -26.90 |
| Energy contribution | -27.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9501619 97 - 20766785 CCGCAGCGCGAGAUGCUCCUGGGCACCUGACGUCAACUGCCGCCCGCGUCCGUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCAC------------------AG-- ..((((((((((.((((....)))).))..)))....(((((((((((.....))))).)).))))))))).(((............))).....------------------..-- ( -35.40) >DroVir_CAF1 2249 93 - 1 CCGCAACGCGAGCUGCUGCUGGGCACCUGACGUCAGCUGCCGCCGGCAUCCAACGCGAUGUCCGCAGCGGCAGCCAAUAGCCACAC------AUC------------------AGCA .(((...))).((((.((.(((((...((....))(((((((((((((((......)))).)))..)))))))).....))).)))------).)------------------))). ( -39.70) >DroPse_CAF1 2065 88 - 1 CCACAGCGAGAGAUGCUUUUGGGCACCUGACGUCAGCUGCCGCCGGCGUCUAGCUCGACGCUGGCAGCUGCAGAAAAUCA---------UACCAC------------------AA-- ...((((..............((((.(((....))).))))(((((((((......))))))))).))))..........---------......------------------..-- ( -34.50) >DroEre_CAF1 2398 97 - 1 CCGCAGCGCGAGAUGCUCUUGGGCACCUGACGUCAACUGCCGCCCGCGUCCAUUGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACUCUCACCAC------------------AG-- ..((((((((((.((((....)))).))..)))....(((((((((((.....))))).)).))))))))).((((..........)))).....------------------..-- ( -36.00) >DroYak_CAF1 2030 97 - 1 CCGCAGCGCGAGAUUCUCCUGGGCACCUGACGUCAGCUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCAC------------------AG-- .(((...))).(((((((..((....))...(.(((((((((((((((.....))))).)).))))))))).)))))))................------------------..-- ( -39.90) >DroAna_CAF1 2098 115 - 1 CCGCAGCGGGAAAUGCUGCUGGGCACCUGACGUCAACUGCCGCCUGCGACUUCCACGCCGCAGGCAGCUGCAGAAAACCGCCACACACACACCACAGGAUCAGUCACACACACAG-- .....((((....(((.(((.((((..((....))..))))(((((((.(......).)))))))))).))).....))))..................................-- ( -36.30) >consensus CCGCAGCGCGAGAUGCUCCUGGGCACCUGACGUCAACUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCAC__________________AG__ ..(((((..............((((..((....))..))))(((.(((((......))))).))).))))).............................................. (-26.90 = -27.15 + 0.25)

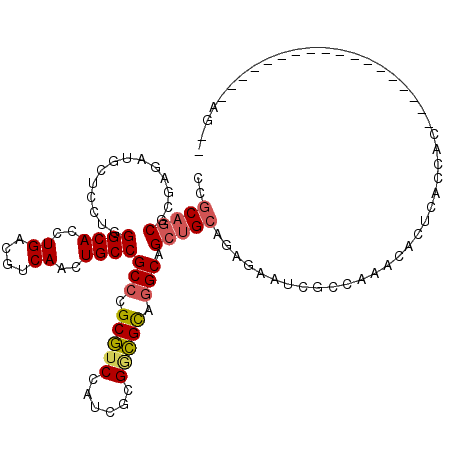
| Location | 9,501,636 – 9,501,756 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -50.53 |
| Consensus MFE | -38.36 |
| Energy contribution | -38.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
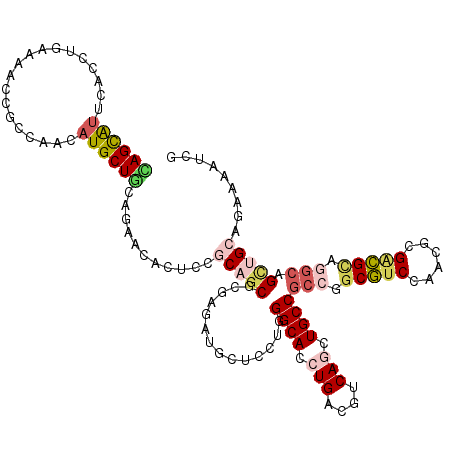
>2R_DroMel_CAF1 9501636 120 + 20766785 CGAUUCUCUGCAGCUGCCUGCGCCGCGACGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUCCUCAGGUGAAUGCUA ..((((((((..(((((((((((((...))).)))))))))).((..((((.((((((......)))))).))(((((((......)))))))..))..))......)))).)))).... ( -58.00) >DroVir_CAF1 2262 120 + 1 CUAUUGGCUGCCGCUGCGGACAUCGCGUUGGAUGCCGGCGGCAGCUGACGUCAGGUGCCCAGCAGCAGCUCGCGUUGCGGAGCAUUUUGCAGCAUAUUGGCGGUUUUUAAAUGAAUGCUG .((..((((((((((((((.((((......)))))).((((((.(((....))).))))..)).)))))....((((((((....)))))))).....)))))))..))........... ( -48.50) >DroPse_CAF1 2073 120 + 1 UGAUUUUCUGCAGCUGCCAGCGUCGAGCUAGACGCCGGCGGCAGCUGACGUCAGGUGCCCAAAAGCAUCUCUCGCUGUGGCGUGCUCUGAAGCAUAUUGGCGGUUUUCAGGUGAAUACUG (((....((((.((((((.(((((......))))).)))))).(((....(((((..((((..(((.......))).))).)..).)))))))......))))...)))........... ( -45.70) >DroGri_CAF1 2213 114 + 1 CUGUUGUUUGCUAC------CGUCGCAUUCGACGACGGCGGCAGCUGACGUCAGGUGCCUAGUAGCAGUUCGCGCUGUGGUGCAUUUUGCAGCAAAUUGGCGGUUUUCAAAUGAACGCUA ..((((((((..((------((((((((..((((.((((....)))).))))..))))...((.((((...((((....))))...)))).)).....))))))...))))).))).... ( -43.00) >DroYak_CAF1 2047 120 + 1 CGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGCUGACGUCAGGUGCCCAGGAGAAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUCCUCAGGUGAAUACUG .((((((((.((((((((.((.(((((.....)))))))))))))))......((...)).))))))))((((.(((.(((.((((..((.....)).)))).))).)))))))...... ( -55.40) >DroAna_CAF1 2133 120 + 1 CGGUUUUCUGCAGCUGCCUGCGGCGUGGAAGUCGCAGGCGGCAGUUGACGUCAGGUGCCCAGCAGCAUUUCCCGCUGCGGAGUGUUCUGUAGCAUGUUGGCGGUUUUCAGGUGAAUACUA .(((.(((.((.((((((((((((......)))))))))))).((..(((..((((((......))))))..)(((((((......)))))))..))..)).........))))).))). ( -52.60) >consensus CGAUUCUCUGCAGCUGCCUGCGUCGCGAUGGACGCCGGCGGCAGCUGACGUCAGGUGCCCAGCAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUAUUGGCGGUUUUCAGGUGAAUACUA ..((((.((((.((((((((((((......)))))))))))).))...((((((((((......)))))....(((((((......)))))))....))))).......)).)))).... (-38.36 = -38.82 + 0.45)
| Location | 9,501,636 – 9,501,756 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -26.19 |
| Energy contribution | -27.36 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9501636 120 - 20766785 UAGCAUUCACCUGAGGACCGCCAACAUGCUGCAGAACACUCCGCAGCGCGAGAUGCUCCUGGGCACCUGACGUCAACUGCCGCCCGCGUCCGUCGCGGCGCAGGCAGCUGCAGAGAAUCG ..(.((((..(((((((((((......(((((.((....)).))))))))((.((((....)))).))...)))..((((((((((((.....))))).)).))))))).))).))))). ( -44.00) >DroVir_CAF1 2262 120 - 1 CAGCAUUCAUUUAAAAACCGCCAAUAUGCUGCAAAAUGCUCCGCAACGCGAGCUGCUGCUGGGCACCUGACGUCAGCUGCCGCCGGCAUCCAACGCGAUGUCCGCAGCGGCAGCCAAUAG ((((((...................))))))..........(((...))).((((((((((((((.(((....))).))))..(((((((......)))).)))))))))))))...... ( -43.51) >DroPse_CAF1 2073 120 - 1 CAGUAUUCACCUGAAAACCGCCAAUAUGCUUCAGAGCACGCCACAGCGAGAGAUGCUUUUGGGCACCUGACGUCAGCUGCCGCCGGCGUCUAGCUCGACGCUGGCAGCUGCAGAAAAUCA .....(((..(((((....((......))))))).((((((....)))......(((....((((.(((....))).))))(((((((((......))))))))))))))).)))..... ( -41.20) >DroGri_CAF1 2213 114 - 1 UAGCGUUCAUUUGAAAACCGCCAAUUUGCUGCAAAAUGCACCACAGCGCGAACUGCUACUAGGCACCUGACGUCAGCUGCCGCCGUCGUCGAAUGCGACG------GUAGCAAACAACAG ..(((((...(((........)))..((.(((.....))).)).)))))....((((....((((.(((....))).))))((((((((.....))))))------))))))........ ( -34.90) >DroYak_CAF1 2047 120 - 1 CAGUAUUCACCUGAGGACCGCCAACAUGCUGCAGAACACUCCGCAGCGCGAGAUUCUCCUGGGCACCUGACGUCAGCUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCG ..(.((((.((.(((((((((......(((((.((....)).)))))))).).)))))..))....(((....(((((((((((((((.....))))).)).))))))))))).))))). ( -49.90) >DroAna_CAF1 2133 120 - 1 UAGUAUUCACCUGAAAACCGCCAACAUGCUACAGAACACUCCGCAGCGGGAAAUGCUGCUGGGCACCUGACGUCAACUGCCGCCUGCGACUUCCACGCCGCAGGCAGCUGCAGAAAACCG ..((.(((.........((((.....(((.............)))))))....(((.(((.((((..((....))..))))(((((((.(......).)))))))))).)))))).)).. ( -36.82) >consensus CAGCAUUCACCUGAAAACCGCCAACAUGCUGCAGAACACUCCGCAGCGCGAGAUGCUCCUGGGCACCUGACGUCAGCUGCCGCCGGCGUCCAACGCGACGCAGGCAGCUGCAGAAAAUCG ((((((...................))))))...........(((((..............((((.(((....))).))))(((.(((((......))))).))).)))))......... (-26.19 = -27.36 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:34 2006