
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,493,593 – 9,493,747 |
| Length | 154 |
| Max. P | 0.983520 |

| Location | 9,493,593 – 9,493,707 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -24.49 |
| Energy contribution | -25.85 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
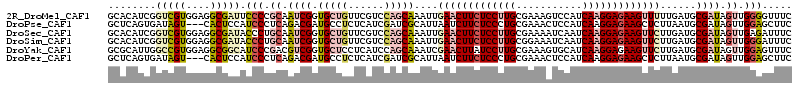
>2R_DroMel_CAF1 9493593 114 + 20766785 GCACAUCGGUCGUGGAGGCGAUUCCCCGCAAUCGGUGCUGUUCGUCCAGCAAAUUGAACUUCUCCUUGCGAAAGUCCAUCAAGGAGAAGUUUUUGAUGCGAUAGUUGGGGUUUC ((.(((.....)))...))((..((((((.((((.(((((......)))))....((((((((((((((....).....)))))))))))))......)))).).)))))..)) ( -40.90) >DroPse_CAF1 5940 111 + 1 GCUCAGUGAUAGU---CACUCCAUCCCUCAGACGAUGCCUCUCAUCGAUCGCAUUAAUCUUCUCCCUGCGAAACUCCAUCAAGGAGAAGCUCUUAAUGCGAUAGUUGGAGCUUC ((((((((.....---))))........(((.(((((.....)))))((((((((((.(((((((.((...........)).)))))))...))))))))))..)))))))... ( -34.40) >DroSec_CAF1 5950 114 + 1 GCACAUCGGUCGUGGAGGCGAUACCCUGCAAUCGGUGCUGUUCGUCCAGCAAAUUGAACUUCUCCUUGCGAAAAUCAAUCAAGGAGAAGUUCUUGAUGCGAUAGUUGAGAUUUC ((((((((((.(..(.((.....)))..).))))))).))).......(((....(((((((((((((...........)))))))))))))....)))............... ( -37.30) >DroSim_CAF1 6020 114 + 1 GCACAUCGGUCGUGGAGGCGAUACCCUGCAAUCGGUGCUGUUCGUCCAGCAAAUUGAACUUCUCCUUGCGGAAAUCAAUCAAGGAGAAGUUCUUGAUGCGAUAGUUGGGAUUUC ........(((((....))))).(((.((.((((.(((((......)))))....(((((((((((((..(....)...)))))))))))))......)))).)).)))..... ( -41.40) >DroYak_CAF1 6230 114 + 1 GCGCAUUGGCCGUGGAGGCGGCAUCCCGACGUCGGUGCUCCUCAUCCAGCAAAUCGAACUUAUCCUUGCGAAAGUGCAUCAAGGAGAAGUUCUUGAUGCGAUAGUUGGAGUUUC .(((.......)))((((.((((..(((....))))))))))).((((((..(((............((....))((((((((((....))))))))))))).))))))..... ( -39.60) >DroPer_CAF1 5923 111 + 1 GCUCAGUGAUAGU---CACUCCAUCCCUCAGACGAUGCCUCUCAUCGAUCGCAUUAAUCUUCUCCCUGCGAAACUCCAUCAAGGAGAAGCUCUUAAUGCGAUAGUUGGAGCUUC ((((((((.....---))))........(((.(((((.....)))))((((((((((.(((((((.((...........)).)))))))...))))))))))..)))))))... ( -34.40) >consensus GCACAUCGGUCGUGGAGGCGACACCCCGCAAUCGGUGCUGCUCAUCCAGCAAAUUGAACUUCUCCUUGCGAAAAUCCAUCAAGGAGAAGUUCUUGAUGCGAUAGUUGGAGUUUC ........(((((....))))).(((.((.((((.(((((......)))))....(((((((((((((...........)))))))))))))......)))).)).)))..... (-24.49 = -25.85 + 1.36)
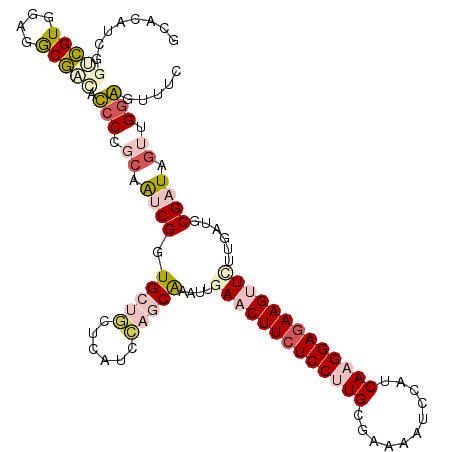
| Location | 9,493,593 – 9,493,707 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9493593 114 - 20766785 GAAACCCCAACUAUCGCAUCAAAAACUUCUCCUUGAUGGACUUUCGCAAGGAGAAGUUCAAUUUGCUGGACGAACAGCACCGAUUGCGGGGAAUCGCCUCCACGACCGAUGUGC ..............((((((...((((((((((((.(((....))))))))))))))).....(((((......)))))..(.(((.((((......)))).))).))))))). ( -38.50) >DroPse_CAF1 5940 111 - 1 GAAGCUCCAACUAUCGCAUUAAGAGCUUCUCCUUGAUGGAGUUUCGCAGGGAGAAGAUUAAUGCGAUCGAUGAGAGGCAUCGUCUGAGGGAUGGAGUG---ACUAUCACUGAGC ...((((...((((((((((((...((((((((((.(((....))))))))))))).)))))))((.(((((.....))))))).....)))))((((---.....)))))))) ( -43.90) >DroSec_CAF1 5950 114 - 1 GAAAUCUCAACUAUCGCAUCAAGAACUUCUCCUUGAUUGAUUUUCGCAAGGAGAAGUUCAAUUUGCUGGACGAACAGCACCGAUUGCAGGGUAUCGCCUCCACGACCGAUGUGC ..............((((((..(((((((((((((...(.....).)))))))))))))....(((((......))))).((..((.(((......))).))))...)))))). ( -37.40) >DroSim_CAF1 6020 114 - 1 GAAAUCCCAACUAUCGCAUCAAGAACUUCUCCUUGAUUGAUUUCCGCAAGGAGAAGUUCAAUUUGCUGGACGAACAGCACCGAUUGCAGGGUAUCGCCUCCACGACCGAUGUGC ..............((((((..(((((((((((((...(....)..)))))))))))))....(((((......))))).((..((.(((......))).))))...)))))). ( -37.80) >DroYak_CAF1 6230 114 - 1 GAAACUCCAACUAUCGCAUCAAGAACUUCUCCUUGAUGCACUUUCGCAAGGAUAAGUUCGAUUUGCUGGAUGAGGAGCACCGACGUCGGGAUGCCGCCUCCACGGCCAAUGCGC ..............(((((...((((((.((((((.((......)))))))).)))))).....((((...((((.((((((....)))..)))..))))..))))..))))). ( -36.60) >DroPer_CAF1 5923 111 - 1 GAAGCUCCAACUAUCGCAUUAAGAGCUUCUCCUUGAUGGAGUUUCGCAGGGAGAAGAUUAAUGCGAUCGAUGAGAGGCAUCGUCUGAGGGAUGGAGUG---ACUAUCACUGAGC ...((((...((((((((((((...((((((((((.(((....))))))))))))).)))))))((.(((((.....))))))).....)))))((((---.....)))))))) ( -43.90) >consensus GAAACCCCAACUAUCGCAUCAAGAACUUCUCCUUGAUGGACUUUCGCAAGGAGAAGUUCAAUUUGCUGGACGAACAGCACCGACUGCGGGAUAUCGCCUCCACGACCAAUGUGC .....(((.....(((......(((((((((((((.(((....)))))))))))))))).....(((........)))..))).....)))..(((......)))......... (-21.78 = -21.98 + 0.20)
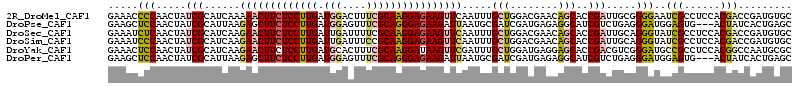
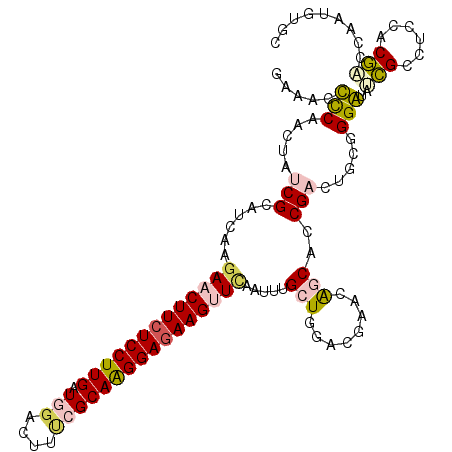
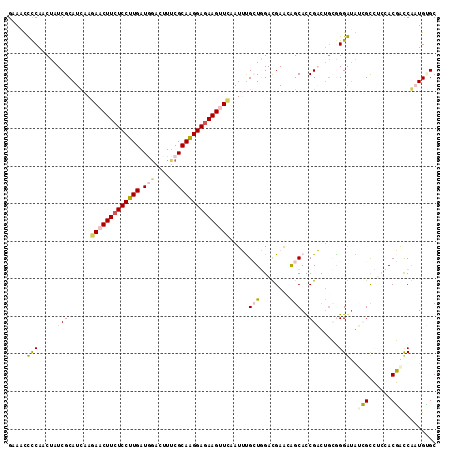
| Location | 9,493,627 – 9,493,747 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9493627 120 - 20766785 CUGCUGCAUAGGACGAACUUUCUAAUCAACACGCCUCGUGGAAACCCCAACUAUCGCAUCAAAAACUUCUCCUUGAUGGACUUUCGCAAGGAGAAGUUCAAUUUGCUGGACGAACAGCAC .(((((..(((((......))))).......((((...(((.....)))......(((.....((((((((((((.(((....))))))))))))))).....))).)).))..))))). ( -34.50) >DroVir_CAF1 8166 120 - 1 CUAUUGCAGCGUACAAACUUCUUGAUGAACACACCGCGUGGCGACUCCAAUUACCGCAUCAAGAGUCUAGCUCUGAUGGAGUUCCGCAAGGAGAAGUAUAAUGCAAUUGACGAAAAGCAG (.((((((..((((...(((((((..((((...((..((((..(......)..))))((((.(((.....))))))))).))))..)))))))..))))..)))))).)........... ( -33.20) >DroPse_CAF1 5971 120 - 1 CUGAUGCAACGAACCAACUUCCUGGUGAACACGCCGCGGGGAAGCUCCAACUAUCGCAUUAAGAGCUUCUCCUUGAUGGAGUUUCGCAGGGAGAAGAUUAAUGCGAUCGAUGAGAGGCAU ...((((..........((((((.(((.......))).))))))(((((.(.((((((((((...((((((((((.(((....))))))))))))).)))))))))).).)).))))))) ( -49.90) >DroYak_CAF1 6264 120 - 1 CUGCUGCACAGGACGAACUUUCUAAUCAACACGCCGCGUGGAAACUCCAACUAUCGCAUCAAGAACUUCUCCUUGAUGCACUUUCGCAAGGAUAAGUUCGAUUUGCUGGAUGAGGAGCAC .((((.(.(((.(((((((..........((((...))))...........((((((((((((........)))))))).....(....)))))))))))...).))).....).)))). ( -30.70) >DroAna_CAF1 6272 120 - 1 UUGUUGCAUCGGACAAACUUUUUGAUCAAUACGCCGCGGGGGAACCCUAACUAUCGCAUCAAGAACUUUGCCUUGAUGGAGUUCCGUAAGGAGAAGUUCAAUGCGAUCGAUGAGAAGCAU .((((.((((((.........................(((....)))......((((((...((((((..((((.((((....))))))))..)))))).))))))))))))...)))). ( -39.80) >DroPer_CAF1 5954 120 - 1 CUGAUGCAACGAACCAACUUCCUGGUGAACACGCCGCGGGGAAGCUCCAACUAUCGCAUUAAGAGCUUCUCCUUGAUGGAGUUUCGCAGGGAGAAGAUUAAUGCGAUCGAUGAGAGGCAU ...((((..........((((((.(((.......))).))))))(((((.(.((((((((((...((((((((((.(((....))))))))))))).)))))))))).).)).))))))) ( -49.90) >consensus CUGAUGCAACGGACAAACUUCCUGAUCAACACGCCGCGGGGAAACUCCAACUAUCGCAUCAAGAACUUCUCCUUGAUGGAGUUUCGCAAGGAGAAGUUCAAUGCGAUCGAUGAGAAGCAU ....(((................................((.....))......(((((...(((((((((((((.(((....)))))))))))))))).)))))...........))). (-19.54 = -20.43 + 0.89)

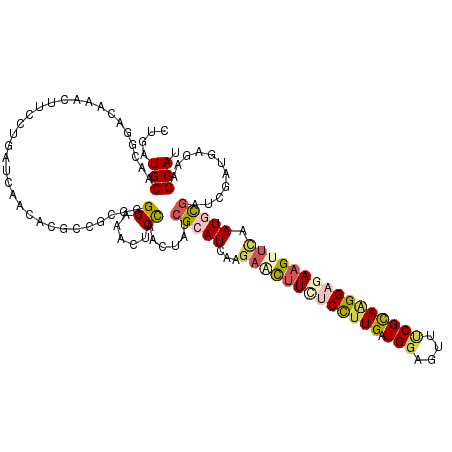
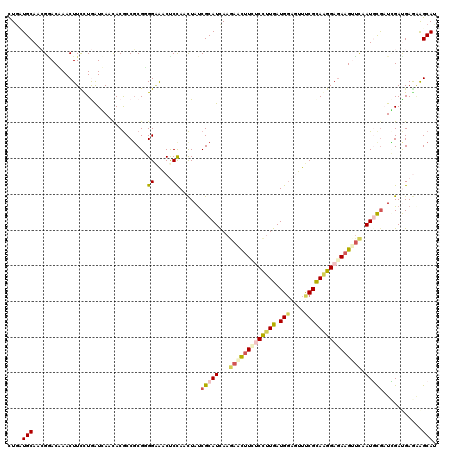
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:27 2006