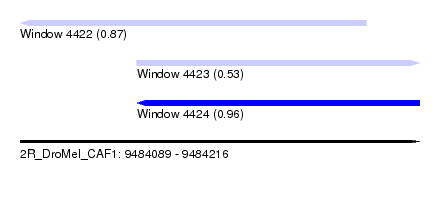
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,484,089 – 9,484,216 |
| Length | 127 |
| Max. P | 0.961526 |

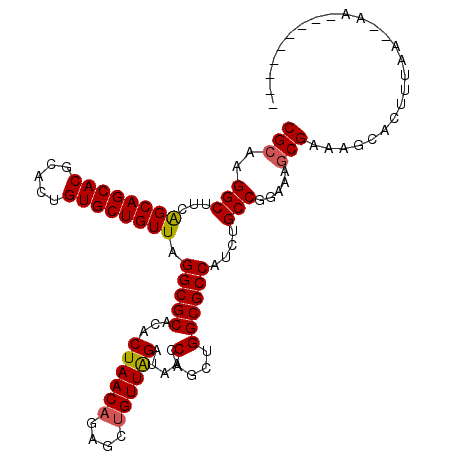
| Location | 9,484,089 – 9,484,199 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -31.56 |
| Energy contribution | -31.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9484089 110 - 20766785 CGCAAGGCUUCAGCAGCACGGACUGUGCUGUUAGGCGCACACUAACAGAGCUGUUGGAUAACCACCUGGCGCCAUCUGCCGGAAAGCGAAAGUGU-UGAA--CAGAAAUUGUA (....)((((.((((((((.....))))))))))))........((((..(((((.(((......((((((.....))))))...((....))))-).))--)))...)))). ( -36.60) >DroSim_CAF1 5947 102 - 1 CGCAAGGCUUCGGCAGCACGGACUGUGCUGUUAGGCGCACACUAACAGAGCUGUUAGAUAACCAGCUGGCGCCAUCUGCCGGAAAGCGAAAGCACUUUUA--CA--------- .....(((...((((((((.....)))))))).(((((...((((((....)))))).....(....))))))....))).....((....)).......--..--------- ( -36.10) >DroEre_CAF1 5773 104 - 1 CGCAAGGCUUCAGCAGCACACACUGUGCUGUUAGGCGCAUACUAACAGAGCCGUUGGCCCACCUGUUGGCGCCAUCUGCCGGAAAGCGAGUGCACUUUAAGCAA--------- (((..(((...((((((((.....)))))))).(((((....((((((.(((...)))....)))))))))))....))).....)))..(((.......))).--------- ( -37.20) >DroYak_CAF1 5899 102 - 1 CGCAAGGCUUCAGCAGCACGCACUGUGCUGUUAGGCGCGCUCUAACAGAGCUGUUAGAACGCCUGCUGGCGCCAUCUGCCGGAAAACGAGGGCCCUUUAA--AA--------- .....((((((((((((((.....))))))))(((((...(((((((....))))))).))))).((((((.....)))))).....)).))))......--..--------- ( -39.30) >consensus CGCAAGGCUUCAGCAGCACGCACUGUGCUGUUAGGCGCACACUAACAGAGCUGUUAGAUAACCAGCUGGCGCCAUCUGCCGGAAAGCGAAAGCACUUUAA__AA_________ (((..(((...((((((((.....)))))))).(((((...((((((....)))))).....(....))))))....))).....)))......................... (-31.56 = -31.62 + 0.06)

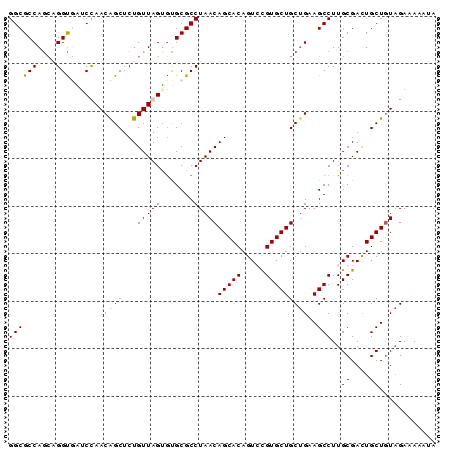
| Location | 9,484,126 – 9,484,216 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -22.29 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9484126 90 + 20766785 GGCGCCAGGUGGUUAUCCAACAGCUCUGUUAGUGUGCGCCUAACAGCACAGUCCGUGCUGCUGAAGCCUUGCGACUGCUGUAGAAAAUUA (((((.((((((....))..((((...(((((.(....))))))(((((.....)))))))))..)))).))).)).............. ( -27.60) >DroSim_CAF1 5976 90 + 1 GGCGCCAGCUGGUUAUCUAACAGCUCUGUUAGUGUGCGCCUAACAGCACAGUCCGUGCUGCCGAAGCCUUGCGACUGCUGUUGAAAAUUA (((((((.........((((((....)))))))).)))))...((((((((((((.(((.....)))..)).))))).)))))....... ( -30.40) >DroEre_CAF1 5804 90 + 1 GGCGCCAACAGGUGGGCCAACGGCUCUGUUAGUAUGCGCCUAACAGCACAGUGUGUGCUGCUGAAGCCUUGCGAUUGCUCUAGAGAAAUA (((((..((..(..((((...))).)..)..))..)))))...((((((.....))))))(((.(((.........))).)))....... ( -28.20) >DroYak_CAF1 5928 90 + 1 GGCGCCAGCAGGCGUUCUAACAGCUCUGUUAGAGCGCGCCUAACAGCACAGUGCGUGCUGCUGAAGCCUUGCGAUUGCUGUAGAAAAAUA ((((((....)))(((((((((....)))))))))..)))..((((((...((((.(((.....)))..))))..))))))......... ( -36.80) >consensus GGCGCCAGCAGGUGAUCCAACAGCUCUGUUAGUGUGCGCCUAACAGCACAGUCCGUGCUGCUGAAGCCUUGCGACUGCUGUAGAAAAAUA ((((((....))).......((((...(((((.......)))))(((((.....)))))))))..)))..((....))............ (-22.29 = -22.22 + -0.06)

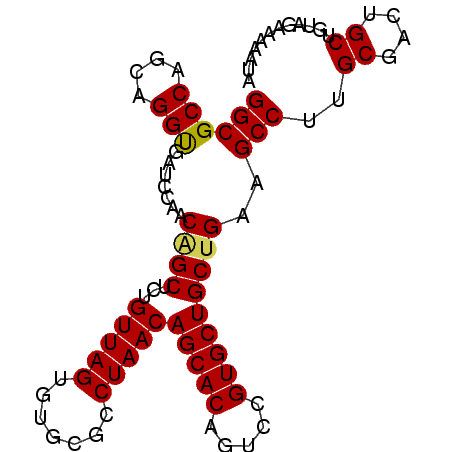
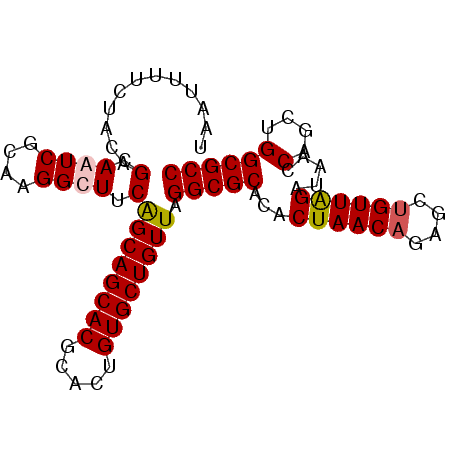
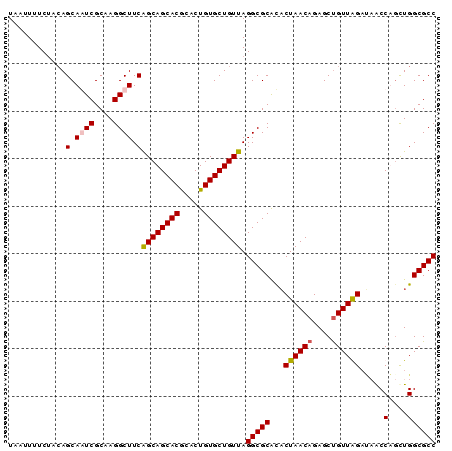
| Location | 9,484,126 – 9,484,216 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9484126 90 - 20766785 UAAUUUUCUACAGCAGUCGCAAGGCUUCAGCAGCACGGACUGUGCUGUUAGGCGCACACUAACAGAGCUGUUGGAUAACCACCUGGCGCC ............(.((((....)))).)((((((((.....)))))))).(((((...((((((....)))))).....(....)))))) ( -30.60) >DroSim_CAF1 5976 90 - 1 UAAUUUUCAACAGCAGUCGCAAGGCUUCGGCAGCACGGACUGUGCUGUUAGGCGCACACUAACAGAGCUGUUAGAUAACCAGCUGGCGCC ............(.((((....)))).)((((((((.....)))))))).(((((...((((((....)))))).....(....)))))) ( -30.80) >DroEre_CAF1 5804 90 - 1 UAUUUCUCUAGAGCAAUCGCAAGGCUUCAGCAGCACACACUGUGCUGUUAGGCGCAUACUAACAGAGCCGUUGGCCCACCUGUUGGCGCC ..........((((...(....))))).((((((((.....)))))))).(((((....((((((.(((...)))....))))))))))) ( -29.40) >DroYak_CAF1 5928 90 - 1 UAUUUUUCUACAGCAAUCGCAAGGCUUCAGCAGCACGCACUGUGCUGUUAGGCGCGCUCUAACAGAGCUGUUAGAACGCCUGCUGGCGCC ..........(((((..(....)((((.((((((((.....))))))))))))(((.(((((((....))))))).))).)))))..... ( -36.10) >consensus UAAUUUUCUACAGCAAUCGCAAGGCUUCAGCAGCACGCACUGUGCUGUUAGGCGCACACUAACAGAGCUGUUAGAUAACCAGCUGGCGCC ............(.((((....)))).)((((((((.....)))))))).(((((...((((((....)))))).....(....)))))) (-27.29 = -27.60 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:23 2006