| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,471,423 – 9,471,536 |
| Length | 113 |
| Max. P | 0.940358 |

| Location | 9,471,423 – 9,471,536 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.88 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
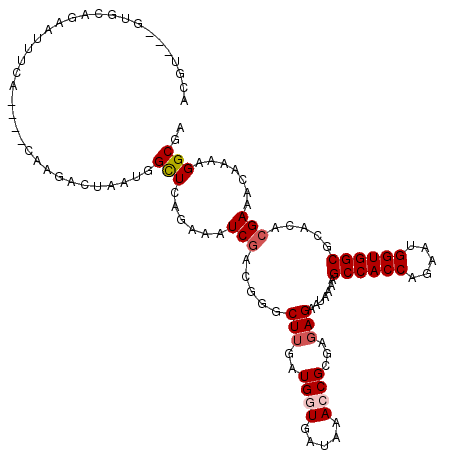
>2R_DroMel_CAF1 9471423 113 - 20766785 ACGU---GUGCGGAAUUGAA----CAAUACAAUUGGCUUAGAAAUCGACGGGCUUGAUGGUGAUGAACCGGGAGAGAAUGAAGGCCACCAGAAUGGUGGCGCACACGAAACAGAAGGCGA .(((---((((..(((((..----.....)))))..............((..(((..((((.....))))...)))..))...((((((.....)))))))))))))............. ( -34.10) >DroPse_CAF1 7050 117 - 1 ACGU---GUGCAGAAUUUCAAUUUCGGGACUAACAGUUCAGAAGUCGACUGGCUUGAUGGUGAUAAACCGCGAGAGAAUAAAAGCCACCAGUAUGGUGGCACAGACGAAGCAAAAGGCGA .(((---.(((..........((((.((((.....)))).))))(((.(((.((((.((((.....)))))))).........((((((.....)))))).))).))).)))....))). ( -36.50) >DroGri_CAF1 8275 99 - 1 --UG---GUUCAG------U----------UAAUGGUUUAGAAAUCGACGGGCUUGAUGGUGAUAAAGCGCGACAGGAUAAAUGCCACCAGAACGGUGGCGCAAACGAAACAAAAGGCGA --..---(((..(------(----------(....(((((....((..((.((((..........)))).))...)).)))))((((((.....))))))...)))..)))......... ( -22.30) >DroEre_CAF1 7522 113 - 1 ACGU---GUGCGGAAUUAGA----CAAGACAUUUUGCUCAGAAAUCGACGGGCUUGAUGGUGAUGAACCGGGAGAGAAUGAAUGCCACCAGAAUGGUGGCGCACACGAAACAGAAGGCGA .(((---((((.........----.....((((((.(((....(((((.....)))))(((.....)))..)))))))))...((((((.....)))))))))))))............. ( -37.70) >DroAna_CAF1 7288 115 - 1 ACGUGGUGUGCAGAAUUUGA----CAA-ACUUUUGGCUUAAAAGUCAACGGGCUUAAUGGUGAUAAAACGGGAGAGAAUAAAGGCCACCAGAAAGGUGGCGCACACGAAGCAAAAGGCAA .....((((((.........----...-.((.((((((....)))))).)).(((..(.((......)).)..))).......((((((.....))))))))))))...((.....)).. ( -32.10) >DroPer_CAF1 6988 117 - 1 ACGU---GUGCAGAAUUUCAAUUUCGGGACUAACAGUUCAGAAGUCGACUGGCUUGAUGGUGAUAAACCGCGAGAGAAUAAAAGCCACCAGUAUGGUGGCACAGACGAAGCAAAAGGCGA .(((---.(((..........((((.((((.....)))).))))(((.(((.((((.((((.....)))))))).........((((((.....)))))).))).))).)))....))). ( -36.50) >consensus ACGU___GUGCAGAAUUUCA____CAAGACUAAUGGCUCAGAAAUCGACGGGCUUGAUGGUGAUAAACCGCGAGAGAAUAAAAGCCACCAGAAUGGUGGCGCACACGAAACAAAAGGCGA ...................................(((......(((.....(((..((((.....))))...))).......((((((.....)))))).....))).......))).. (-17.80 = -18.22 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:21 2006