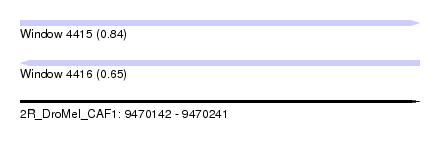
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,470,142 – 9,470,241 |
| Length | 99 |
| Max. P | 0.841488 |

| Location | 9,470,142 – 9,470,241 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
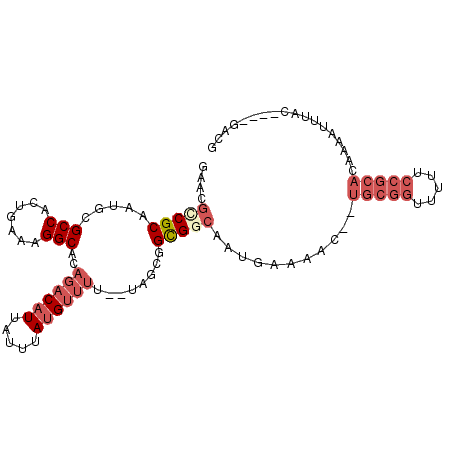
>2R_DroMel_CAF1 9470142 99 + 20766785 CGUC----GUAAAUUUUGUGCGGAAAAACCGCAGAGUUUUCAUUGCCGCCGUGAAAAAAACAUAAAUAAUGUCUGUGCCUUUCAGUGGCGCAUUGCGGCGUUC .((.----((.((..((.(((((.....))))).))..)).)).))(((((..(.....((((.....)))).((((((.......)))))))..)))))... ( -31.10) >DroPse_CAF1 5910 88 + 1 CGUCGCUCGUAAUUAUUGUU------AAUUCUG--UUUUGCAAUUCCAC---UA--AUUCCUUAA--AAUGUCGGUGCCAUUUAGUGGCACUCUGCGACGUUC ((((((..((....(((((.------((.....--.)).)))))...))---..--.........--......((((((((...))))))))..))))))... ( -22.10) >DroSec_CAF1 6281 95 + 1 CGUC----GUAAAUUUUGUGCGGAAAAACCGCA--GUUUUCAUUGCCGCCGCUA--AAAACAUAAAUAAUGUCUGUGCCUUUCAGUGGCGCAUUGCGGCGUUC ((((----((((......(((((.....)))))--........(((.(((((((--((..((((.........))))..))).)))))))))))))))))... ( -30.30) >DroSim_CAF1 6286 97 + 1 CGUC----GUGAAUUUUGUGCGGAAAAACCGCAGAGUUUUUAUUGCCGCCGCUA--AAAACAUAAAUAAUGUCUGUGCCUUUCAGUGGCGCAUUGCGGCGUUC .((.----(((((..((.(((((.....))))).))..))))).))((((((..--...((((.....)))).((((((.......))))))..))))))... ( -35.20) >DroEre_CAF1 6262 95 + 1 CGUC----GUAAUUUUUGUGCGGAAAAACCGCA--GUUUUCAUUGGCGCUAAGA--AAAACAUAAAUAAUGUCUGUGCCAUUCAGUGGCGCAUUGCGGCGUUC ((((----(((((.....(((((.....)))))--(((((..(((....)))..--))))).............(((((((...))))))))))))))))... ( -29.70) >DroYak_CAF1 7957 95 + 1 CGUC----GUAACUUUUGUGCGGAAAAACCGCA--GUUUUUAUUGGCGCUAAGA--AAAACAUAAAUAAUGUCUGUGCCUUUCAGUGGCGCAUUGCGGCGUUC ((((----((((......(((((.....)))))--..........((((((.((--((..((((.........))))..))))..)))))).))))))))... ( -29.40) >consensus CGUC____GUAAAUUUUGUGCGGAAAAACCGCA__GUUUUCAUUGCCGCCGAGA__AAAACAUAAAUAAUGUCUGUGCCUUUCAGUGGCGCAUUGCGGCGUUC ((((..............(((((.....))))).....................................((.((((((.......))))))..))))))... (-17.02 = -17.00 + -0.02)

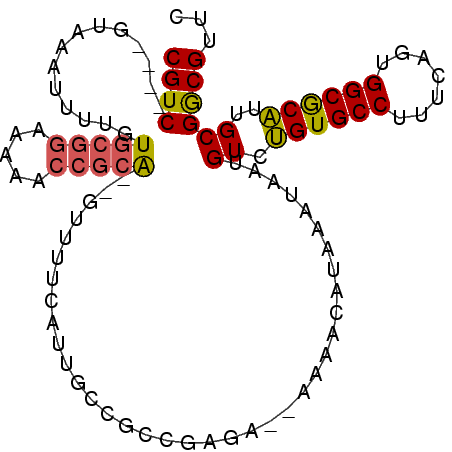
| Location | 9,470,142 – 9,470,241 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -16.45 |
| Energy contribution | -18.23 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9470142 99 - 20766785 GAACGCCGCAAUGCGCCACUGAAAGGCACAGACAUUAUUUAUGUUUUUUUCACGGCGGCAAUGAAAACUCUGCGGUUUUUCCGCACAAAAUUUAC----GACG ....(((((..((.(((.......))).))(((((.....))))).........)))))...........(((((.....)))))..........----.... ( -26.00) >DroPse_CAF1 5910 88 - 1 GAACGUCGCAGAGUGCCACUAAAUGGCACCGACAUU--UUAAGGAAU--UA---GUGGAAUUGCAAAA--CAGAAUU------AACAAUAAUUACGAGCGACG ...((((((.(.((((((.....)))))))......--....(.(((--((---.((......(....--..)....------..)).))))).)..)))))) ( -20.60) >DroSec_CAF1 6281 95 - 1 GAACGCCGCAAUGCGCCACUGAAAGGCACAGACAUUAUUUAUGUUUU--UAGCGGCGGCAAUGAAAAC--UGCGGUUUUUCCGCACAAAAUUUAC----GACG ...((((((..((.(((.......))).))(((((.....)))))..--..))))))...........--(((((.....)))))..........----.... ( -28.40) >DroSim_CAF1 6286 97 - 1 GAACGCCGCAAUGCGCCACUGAAAGGCACAGACAUUAUUUAUGUUUU--UAGCGGCGGCAAUAAAAACUCUGCGGUUUUUCCGCACAAAAUUCAC----GACG (((((((((..((.(((.......))).))(((((.....)))))..--..)))))).............(((((.....))))).....)))..----.... ( -29.10) >DroEre_CAF1 6262 95 - 1 GAACGCCGCAAUGCGCCACUGAAUGGCACAGACAUUAUUUAUGUUUU--UCUUAGCGCCAAUGAAAAC--UGCGGUUUUUCCGCACAAAAAUUAC----GACG ....(.(((..((.((((.....)))).))(((((.....)))))..--.....))).).........--(((((.....)))))..........----.... ( -20.10) >DroYak_CAF1 7957 95 - 1 GAACGCCGCAAUGCGCCACUGAAAGGCACAGACAUUAUUUAUGUUUU--UCUUAGCGCCAAUAAAAAC--UGCGGUUUUUCCGCACAAAAGUUAC----GACG ...((.((.(((((((....((((((((.(((.....))).))))))--))...))))..........--(((((.....))))).....))).)----).)) ( -21.80) >consensus GAACGCCGCAAUGCGCCACUGAAAGGCACAGACAUUAUUUAUGUUUU__UAGCGGCGGCAAUGAAAAC__UGCGGUUUUUCCGCACAAAAUUUAC____GACG ....(((((.....(((.......)))..((((((.....))))))........)))))...........(((((.....))))).................. (-16.45 = -18.23 + 1.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:16 2006