| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
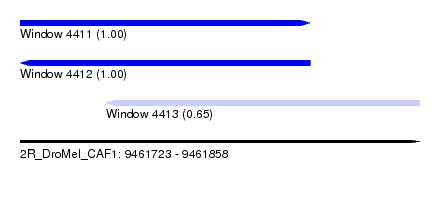
| Location | 9,461,723 – 9,461,858 |
| Length | 135 |
| Max. P | 0.999985 |

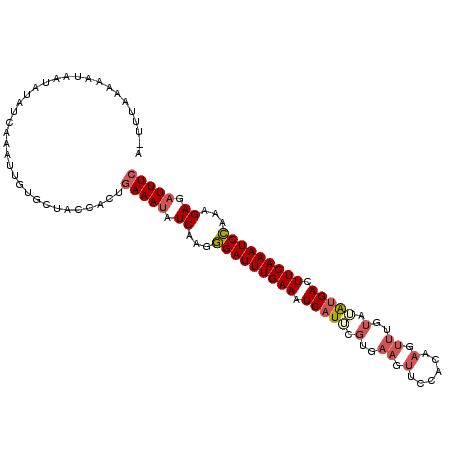
| Location | 9,461,723 – 9,461,821 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.96 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.40 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9461723 98 + 20766785 A-UUUAAAAAUUUUUU----------GCCACCACUGAAAUAUCAAGGGAUUUGAAAUCAUUCGUGAAGUUCCACCAGUUUGUAUAUGAUUUCAAAUCCAAAGAGAUUUC .-..............----------.........(((((.((...((((((((((((((..(..((.(......).))..)..))))))))))))))...)).))))) ( -23.30) >DroSec_CAF1 48528 108 + 1 A-UUUAAAAAUAAUAUAUCAAAUUGUGAUACCACUGAAAUAUCAAGGGAUUUGAAAUCAUUCGUGAAGUUCCAAAAGUUUGUAUAUGACUUCAAAUCCAAAGAGAUUUC .-.............(((((.....))))).....(((((.((...(((((((((.((((..(..((.((....)).))..)..)))).)))))))))...)).))))) ( -22.30) >DroSim_CAF1 58329 99 + 1 A-UUUAAAAAUAAUAUAUCAAAUUGUG---------AAAUAUCAAGGGAUUUGAAAUCAUGCGUGAAGUUGCACAAGUUUGUAUACGACUUCAAAUCCAAAGAUAUUUC .-........................(---------(((((((...(((((((((.((.((((......))))...((......)))).)))))))))...)))))))) ( -23.70) >DroYak_CAF1 49009 98 + 1 GAC-UAAAAAGACAAUAUCAAAUUGUGCUACCACUGAAACAUCAAGUGAUUUGAAAUCAUUC----------ACUAGUUUGUAAGUGACUUCAAAUCGCAUGAGAUUUC ...-.....(((((((.....))))).))......((((..(((.((((((((((.(((((.----------((......)).))))).)))))))))).)))..)))) ( -25.20) >consensus A_UUUAAAAAUAAUAUAUCAAAUUGUGCUACCACUGAAAUAUCAAGGGAUUUGAAAUCAUUCGUGAAGUUCCACAAGUUUGUAUAUGACUUCAAAUCCAAAGAGAUUUC ...................................(((((.((...(((((((((.(((((.(..((.(......).))..).))))).)))))))))...)).))))) (-13.84 = -15.40 + 1.56)

| Location | 9,461,723 – 9,461,821 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.96 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.21 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 5.40 |
| SVM RNA-class probability | 0.999985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9461723 98 - 20766785 GAAAUCUCUUUGGAUUUGAAAUCAUAUACAAACUGGUGGAACUUCACGAAUGAUUUCAAAUCCCUUGAUAUUUCAGUGGUGGC----------AAAAAAUUUUUAAA-U (((((.((...((((((((((((((...(.....)((((....))))..))))))))))))))...)).))))).........----------..............-. ( -24.10) >DroSec_CAF1 48528 108 - 1 GAAAUCUCUUUGGAUUUGAAGUCAUAUACAAACUUUUGGAACUUCACGAAUGAUUUCAAAUCCCUUGAUAUUUCAGUGGUAUCACAAUUUGAUAUAUUAUUUUUAAA-U (((((.((...((((((((((((((..........(((........)))))))))))))))))...)).)))))....((((((.....))))))............-. ( -25.10) >DroSim_CAF1 58329 99 - 1 GAAAUAUCUUUGGAUUUGAAGUCGUAUACAAACUUGUGCAACUUCACGCAUGAUUUCAAAUCCCUUGAUAUUU---------CACAAUUUGAUAUAUUAUUUUUAAA-U ((((((((...((((((((((((((..(((....)))((........))))))))))))))))...)))))))---------)........................-. ( -28.00) >DroYak_CAF1 49009 98 - 1 GAAAUCUCAUGCGAUUUGAAGUCACUUACAAACUAGU----------GAAUGAUUUCAAAUCACUUGAUGUUUCAGUGGUAGCACAAUUUGAUAUUGUCUUUUUA-GUC (((((.(((.(.((((((((((((.((((......))----------)).)))))))))))).).))).))))).(((....)))....................-... ( -27.50) >consensus GAAAUCUCUUUGGAUUUGAAGUCAUAUACAAACUUGUGGAACUUCACGAAUGAUUUCAAAUCCCUUGAUAUUUCAGUGGUAGCACAAUUUGAUAUAUUAUUUUUAAA_U (((((.((...((((((((((((((........................))))))))))))))...)).)))))................................... (-18.02 = -18.21 + 0.19)

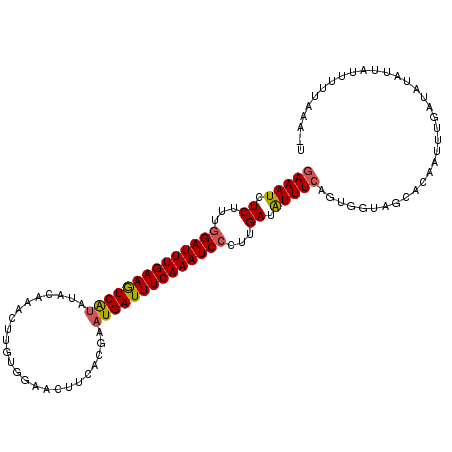

| Location | 9,461,752 – 9,461,858 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -7.58 |
| Energy contribution | -7.74 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9461752 106 - 20766785 CGCACGCGGUCCCUCAAAAUGUUGAAUCGAACUUAUAGAAAUCUCUUUGGAUUUGAAAU-----------CAUAUACAAACUGGUGGAACUUCACGAAUGAUUUCAAAUCCCUUGAU ......((((...((((....))))))))..............((...(((((((((((-----------(((...(.....)((((....))))..))))))))))))))...)). ( -21.50) >DroSec_CAF1 48567 106 - 1 CGCAUGCGGUCCCUCAAAAUGUUGAAUCGAACUCUUAGAAAUCUCUUUGGAUUUGAAGU-----------CAUAUACAAACUUUUGGAACUUCACGAAUGAUUUCAAAUCCCUUGAU ......((((...((((....))))))))..............((...(((((((((((-----------(((..........(((........)))))))))))))))))...)). ( -19.00) >DroSim_CAF1 58359 106 - 1 CGCAUGCGGUCCCUCAAAAUGUUGAAUCGAACUCUUAGAAAUAUCUUUGGAUUUGAAGU-----------CGUAUACAAACUUGUGCAACUUCACGCAUGAUUUCAAAUCCCUUGAU ..((((((((((.((((....))))..........((....)).....)))).((((((-----------.(((((......))))).))))))))))))...((((.....)))). ( -22.60) >DroEre_CAF1 48605 103 - 1 CACA----GUCCCUCAAAAUGUUGAAUCGAACAUUUAGAAAUCUCUUUCGAUUUGAAAUCAGUCAUAAAUCACUUGCGAGCUACU----------GCAUGAUUUCAAAUCACUUGAU ....----.....((.(((((((......))))))).))..........((((((((((((((..((..((......))..))..----------)).))))))))))))....... ( -21.30) >DroYak_CAF1 49048 92 - 1 CACA----GUCCCCCAAAAUGUUGAAUCGAACAUUUAGAAAUCUCAUGCGAUUUGAAGU-----------CACUUACAAACUAGU----------GAAUGAUUUCAAAUCACUUGAU ....----........(((((((......))))))).......(((.(.((((((((((-----------((.((((......))----------)).)))))))))))).).))). ( -24.00) >consensus CGCA_GCGGUCCCUCAAAAUGUUGAAUCGAACUUUUAGAAAUCUCUUUGGAUUUGAAGU___________CAUAUACAAACUAGUG_AACUUCACGAAUGAUUUCAAAUCCCUUGAU ...........................................((...(((((((((((.........................................)))))))))))...)). ( -7.58 = -7.74 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:13 2006