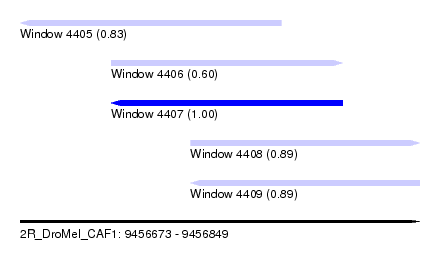
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,456,673 – 9,456,849 |
| Length | 176 |
| Max. P | 0.998805 |

| Location | 9,456,673 – 9,456,788 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -24.81 |
| Energy contribution | -26.78 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
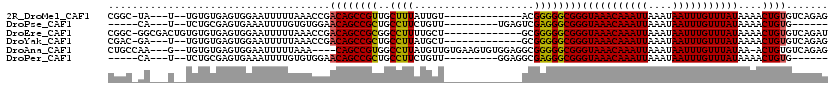
>2R_DroMel_CAF1 9456673 115 - 20766785 UAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG--CCGCAACCCGCUCCCUC---UGACGCAGGUGCGUUUAUCUACAGUCGUACCUGCCUAUCCC ((((((((((....)))))))))).........(((((((...((((....((((..--.))))....)))).)))---))))(((((((((............)))))))))....... ( -38.10) >DroSec_CAF1 43535 115 - 1 UAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG--UCGCAAUCCGCUCCCUC---UGACGCAGGUGCGUUUAUCUACAGUCGUACCUGCCUAACCC ((((((((((....)))))))))).........(((((((...((((....((((..--.))))....)))).)))---))))(((((((((............)))))))))....... ( -38.10) >DroSim_CAF1 52065 115 - 1 UAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG--CCGCAACCCGCUCCCUC---UGACGCAGGUGCGUUUAUCUACAGUCGUACCUGCCUAACCC ((((((((((....)))))))))).........(((((((...((((....((((..--.))))....)))).)))---))))(((((((((............)))))))))....... ( -38.10) >DroEre_CAF1 43684 114 - 1 UAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAUCAAGAGUCAUGUGUGAG---CGCAACCCGCUCCCUC---UGACGCAGGUGCGUUGAUCUACAGUCGUGCCUGCCUAACCC ((((((((((....)))))))))).........((((((....((((..((((....---))))....))))..))---))))(((((..(((((.....))).))..)))))....... ( -36.80) >DroYak_CAF1 43870 117 - 1 UAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGUGAUCCACAACCCGCUCCCUC---UGACGCAGGUGCGUUUAUCUACAGUCGUGCCUGCCUAACCC ((((((((((....)))))))))).........(((((((...((((....((((.....))))....)))).)))---))))(((((..((............))..)))))....... ( -38.80) >DroAna_CAF1 45011 94 - 1 UAAACAAAUUAAAUAAUUUGUUUAUAA-ACUGUGUCAGAGC---------------------CAACCCGCUCCCUCCUAUGACGUAGCGGC----AUCGACAGUCGAACAUCCCUAACCC ((((((((((....))))))))))...-.((((((((((((---------------------......)))).......))))))))((((----.......)))).............. ( -18.61) >consensus UAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG__CCGCAACCCGCUCCCUC___UGACGCAGGUGCGUUUAUCUACAGUCGUACCUGCCUAACCC ((((((((((....)))))))))).........(((((((...((((....((((.....))))....)))).)))...))))(((((((((............)))))))))....... (-24.81 = -26.78 + 1.97)

| Location | 9,456,713 – 9,456,815 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.66 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9456713 102 + 20766785 AGAGGGAGCGGGUUGCGG--CUCACACAUGACUCUUGCUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGUACAAUAAAGCAACGGC ...(((.(((((..((((--.(((....)))...))))...............((((((((((....))))))))))))))).))).................. ( -24.90) >DroSec_CAF1 43575 102 + 1 AGAGGGAGCGGAUUGCGA--CUCACACAUGACUCUUGCUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCACCAUAAGGCAACGGC .(.(((.((((...((((--.(((....)))...))))...............((((((((((....)))))))))).)))).))).).......(....)... ( -24.00) >DroSim_CAF1 52105 102 + 1 AGAGGGAGCGGGUUGCGG--CUCACACAUGACUCUUGCUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCACCAUAAGGCAACGGC ....((.(((((..((((--.........((((..((......)).))))...((((((((((....)))))))))).)))).))))).))....(....)... ( -27.10) >DroEre_CAF1 43724 101 + 1 AGAGGGAGCGGGUUGCG---CUCACACAUGACUCUUGAUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAGCAAAAGGCCGCGGC ...(((.((((((((.(---.(((..((.......))...)))).))).....((((((((((....))))))))))))))))))(((.((.....)).))).. ( -29.20) >DroYak_CAF1 43910 104 + 1 AGAGGGAGCGGGUUGUGGAUCACACACAUGACUCUUGCUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAGCAUAAGGCAGCGGC ...(((.(((((.((((.....))))...........................((((((((((....))))))))))))))))))(((.((.....)).))).. ( -31.30) >consensus AGAGGGAGCGGGUUGCGG__CUCACACAUGACUCUUGCUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCACCAUAAGGCAACGGC ...(((.(((((..((((...(((....)))...))))...............((((((((((....))))))))))))))).)))((........))...... (-23.30 = -23.66 + 0.36)


| Location | 9,456,713 – 9,456,815 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -30.06 |
| Energy contribution | -29.94 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
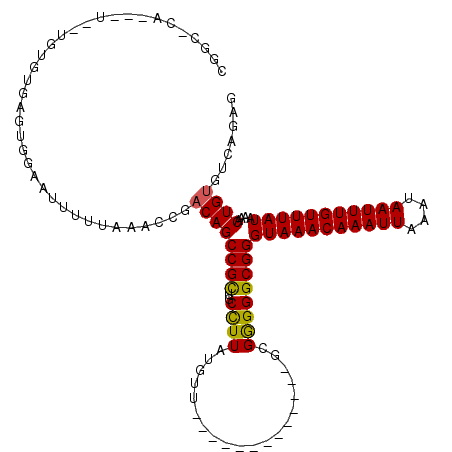
>2R_DroMel_CAF1 9456713 102 - 20766785 GCCGUUGCUUUAUUGUACGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG--CCGCAACCCGCUCCCUCU ...((.((......))))(((((((((((((((((((....))))))))).......(((((((..(((.......))).))).--.))))))))))))))... ( -32.20) >DroSec_CAF1 43575 102 - 1 GCCGUUGCCUUAUGGUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG--UCGCAAUCCGCUCCCUCU (((((......)))))..(((((((((((((((((((....)))))))))))..............((..((.(((....))).--))))...))))))))... ( -34.70) >DroSim_CAF1 52105 102 - 1 GCCGUUGCCUUAUGGUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG--CCGCAACCCGCUCCCUCU (((((......)))))..(((((((((((((((((((....))))))))).......(((((((..(((.......))).))).--.))))))))))))))... ( -35.90) >DroEre_CAF1 43724 101 - 1 GCCGCGGCCUUUUGCUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAUCAAGAGUCAUGUGUGAG---CGCAACCCGCUCCCUCU ...(((((.....)))))(((((((((((((((((((....))))))))).......(((((((.((..........)).))).---))))))))))))))... ( -39.50) >DroYak_CAF1 43910 104 - 1 GCCGCUGCCUUAUGCUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGUGAUCCACAACCCGCUCCCUCU ((.((........)).))(((((((((((((((((((....))))))))).......(((((((..(((.......)))...)))..))))))))))))))... ( -36.30) >consensus GCCGUUGCCUUAUGGUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAGCAAGAGUCAUGUGUGAG__CCGCAACCCGCUCCCUCU ...((.(((....)))))(((((((((((((((((((....))))))))).....(((.((......))..)))....((((.....))))))))))))))... (-30.06 = -29.94 + -0.12)

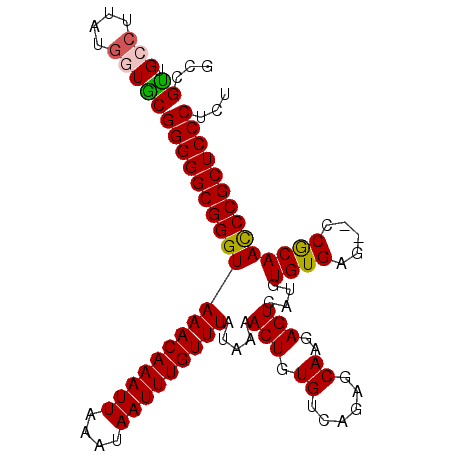
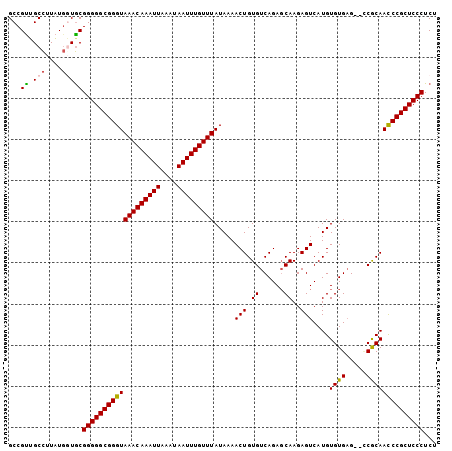
| Location | 9,456,748 – 9,456,849 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.72 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9456748 101 + 20766785 CUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGU-------------ACAAUAAAGCAACGGCUGUCGGUUUAAAAAUUCCACUCACACA--A---UA-GCCG ..........((((((((((((((((....)))))))))...((....)).-------------...)))))))..(((((((.(....................).--)---))-)))) ( -16.85) >DroPse_CAF1 42403 95 + 1 ------CACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCUCGACUCA---------AACAGAAGGCAGCGGCUGUUCCACACAAAAUUUCACUCGCAGA--A---UG----- ------.((((((...((((((((((....))))))))))...(((.((......---------....)).)))...))))))........................--.---..----- ( -16.70) >DroEre_CAF1 43758 106 + 1 AUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGC-------------AGCAAAAGGCCGCGGCUGUCGGUUUAAAAAUUCCACUCACACACAGUCGCC-GCCG ..(((.....((....((((((((((....))))))))))...)).....)-------------)).....(((.((((((((.(....................))))))))).-))). ( -25.65) >DroYak_CAF1 43947 101 + 1 CUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGC-------------AGCAUAAGGCAGCGGCUGUCGGUUUAAAAAUUCCACUCACACA--A---UC-GUCG ....(((.........((((((((((....)))))))))).((((..((((-------------.((.....)).))))..).))).....................--.---..-))). ( -22.30) >DroAna_CAF1 45066 110 + 1 CUCUGACACAGU-UUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCCUCCACACUUCACAACAUAAGGCCACGGCUG----UUUAAAAAUUCCACUCACACA--C---UUGGCAG ..(((.((.(((-...((((((((((....))))))))))...(((...((((.................))))...)))..----....................)--)---))).))) ( -17.23) >DroPer_CAF1 43224 95 + 1 ------CACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCUCGCCUCC---------AACAGAAGGCAGCGGCUGUUCCACACAAAAUUUCACUCGCAGA--A---UG----- ------.((((.....((((((((((....)))))))))).((((....((((.(---------....).)))).))))))))........................--.---..----- ( -19.00) >consensus CUCUGACACAGUUUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGC_____________AACAUAAGGCAGCGGCUGUCGGUUUAAAAAUUCCACUCACACA__A___UC_GCCG .......((((.....((((((((((....)))))))))).((((..((......................))..))))))))..................................... (-11.27 = -11.72 + 0.45)
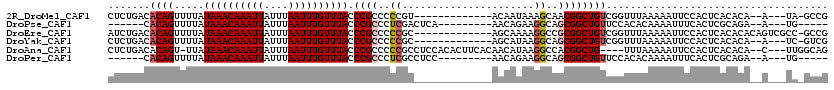

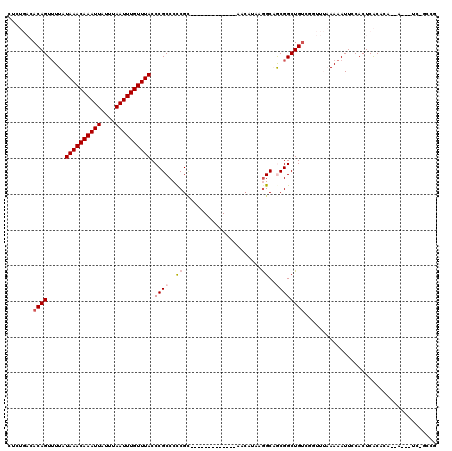
| Location | 9,456,748 – 9,456,849 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -17.22 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9456748 101 - 20766785 CGGC-UA---U--UGUGUGAGUGGAAUUUUUAAACCGACAGCCGUUGCUUUAUUGU-------------ACGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAG ((((-(.---(--((.((...((((...)))).))))).)))))........(((.-------------(((.((....(((((((((((....)))))))))))....)).)))))).. ( -23.90) >DroPse_CAF1 42403 95 - 1 -----CA---U--UCUGCGAGUGAAAUUUUGUGUGGAACAGCCGCUGCCUUCUGUU---------UGAGUCGAGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUG------ -----..---(--((((((((......)))))).)))((((...((((((((....---------......))))))))(((((((((((....)))))))))))....)))).------ ( -25.90) >DroEre_CAF1 43758 106 - 1 CGGC-GGCGACUGUGUGUGAGUGGAAUUUUUAAACCGACAGCCGCGGCCUUUUGCU-------------GCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAU .(((-.(((.((((((.((((.......)))).))..)))).))).))).....((-------------(((.((....(((((((((((....)))))))))))....)).)).))).. ( -32.40) >DroYak_CAF1 43947 101 - 1 CGAC-GA---U--UGUGUGAGUGGAAUUUUUAAACCGACAGCCGCUGCCUUAUGCU-------------GCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAG .(((-(.---.--.((..................(((.(..((((.((.....)).-------------))))..))))(((((((((((....)))))))))))...))..)))).... ( -27.40) >DroAna_CAF1 45066 110 - 1 CUGCCAA---G--UGUGUGAGUGGAAUUUUUAAA----CAGCCGUGGCCUUAUGUUGUGAAGUGUGGAGGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAA-ACUGUGUCAGAG (((((..---.--(((.((((.......)))).)----))(((.(.((((((..((....))..).))))).).)))))(((((((((((....)))))))))))..-.......))).. ( -29.10) >DroPer_CAF1 43224 95 - 1 -----CA---U--UCUGCGAGUGAAAUUUUGUGUGGAACAGCCGCUGCCUUCUGUU---------GGAGGCGAGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUG------ -----..---(--((((((((......)))))).)))(((((((((((((((....---------))))))...)))))(((((((((((....)))))))))))....)))).------ ( -31.20) >consensus CGGC_CA___U__UGUGUGAGUGGAAUUUUUAAACCGACAGCCGCUGCCUUAUGUU_____________GCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAAACUGUGUCAGAG .....................................((((((((..((((....................))))))))(((((((((((....)))))))))))....))))....... (-17.22 = -16.80 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:09 2006