| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,441,642 – 9,441,762 |
| Length | 120 |
| Max. P | 0.991601 |

| Location | 9,441,642 – 9,441,762 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -22.39 |
| Energy contribution | -25.75 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
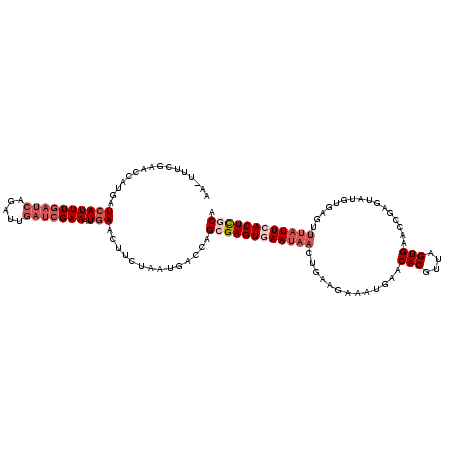
>2R_DroMel_CAF1 9441642 120 + 20766785 UGCGAGUGAAUAAACGCACAUACUCCGUUCACUAACGUGUUCAUUUCUUCAGUUAUUCACUCGCUGGUCGUUAGAAGUUCAUUUCGAUCAAACUGAUCAAAUGAUCAUUGUUCGAAAAGG .((((((((((((.............(..(((....)))..)..........)))))))))))).................((((((.(((..(((((....)))))))).))))))... ( -32.50) >DroSec_CAF1 28893 119 + 1 UGCGAGUGAAUAAACUCACAUACUCGGUUCACUAACGUGUUCAUUUUUUCAGUUAUUCACUCGCUGGUCAUUAGAAGUUCAUUUCGAUCAAUCUGAUCAAAUGAUCAUGGUUCGAAA-UU .((((((((((((.((..........(..(((....)))..)........))))))))))))))((((((((.((((....))))((((.....)))).))))))))..........-.. ( -33.97) >DroSim_CAF1 37133 119 + 1 UGCAAGUGAAUAAACUCACAUACUCGAUUCACUAACGUGUUCAUUUUUUCAGUUAUUCACUCGCUGGUCAUUAGAAGUUCAUUUCGAUCAAUCUGAUCAAAUGAUCAUGGUUCGAAA-UU .((.(((((((((.((.........((..(((....))).))........))))))))))).))((((((((.((((....))))((((.....)))).))))))))..........-.. ( -27.43) >DroEre_CAF1 28973 106 + 1 UGUGAGUGAA----GUUCACUACUCGGUUCACUAACGUGUUCAUUUCUUCAUUUAUUUACUCGCUGGUCAUAAGAAGUUAAUUUCGAUC---------AAAUGAUCUUGAUUCGAAA-UU .(((((((((----............(..(((....)))..).............)))))))))...............((((((((((---------((......)))).))))))-)) ( -23.61) >DroYak_CAF1 29001 119 + 1 UGUGAGUGAAUAAACUCAGCUACAGAGUUCACUAACGUGUUCAUUUCUUUAGUUAUUUACUCGCUGGUCAUUAGAAGUUCAAUUCGAUCAAAUUGAUCAAAUGAUCAUGAUUCUUAA-UG .(((((((((((......((((.((((..(((....))).....)))).)))))))))))))))((((((((.(((......)))((((.....)))).))))))))..........-.. ( -33.00) >consensus UGCGAGUGAAUAAACUCACAUACUCGGUUCACUAACGUGUUCAUUUCUUCAGUUAUUCACUCGCUGGUCAUUAGAAGUUCAUUUCGAUCAAACUGAUCAAAUGAUCAUGGUUCGAAA_UU .((((((((((((................(((....))).............)))))))))))).(((((((.((((....))))((((.....)))).))))))).............. (-22.39 = -25.75 + 3.36)


| Location | 9,441,642 – 9,441,762 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -16.05 |
| Energy contribution | -18.29 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9441642 120 - 20766785 CCUUUUCGAACAAUGAUCAUUUGAUCAGUUUGAUCGAAAUGAACUUCUAACGACCAGCGAGUGAAUAACUGAAGAAAUGAACACGUUAGUGAACGGAGUAUGUGCGUUUAUUCACUCGCA ...((((((.((((((((....)))))..))).))))))......((....))...((((((((((((.((.....(((....((((....))))...)))...)).)))))))))))). ( -34.40) >DroSec_CAF1 28893 119 - 1 AA-UUUCGAACCAUGAUCAUUUGAUCAGAUUGAUCGAAAUGAACUUCUAAUGACCAGCGAGUGAAUAACUGAAAAAAUGAACACGUUAGUGAACCGAGUAUGUGAGUUUAUUCACUCGCA .(-((((((.(..(((((....)))))....).)))))))................((((((((((((((.(.........(((....)))...........).)).)))))))))))). ( -32.15) >DroSim_CAF1 37133 119 - 1 AA-UUUCGAACCAUGAUCAUUUGAUCAGAUUGAUCGAAAUGAACUUCUAAUGACCAGCGAGUGAAUAACUGAAAAAAUGAACACGUUAGUGAAUCGAGUAUGUGAGUUUAUUCACUUGCA .(-((((((.(..(((((....)))))....).)))))))................((((((((((((((.(.....(((.(((....)))..)))......).)).)))))))))))). ( -31.10) >DroEre_CAF1 28973 106 - 1 AA-UUUCGAAUCAAGAUCAUUU---------GAUCGAAAUUAACUUCUUAUGACCAGCGAGUAAAUAAAUGAAGAAAUGAACACGUUAGUGAACCGAGUAGUGAAC----UUCACUCACA ((-((((((.(((((....)))---------))))))))))..((((((((.............))))..)))).......(((....)))....((((((....)----)..))))... ( -17.62) >DroYak_CAF1 29001 119 - 1 CA-UUAAGAAUCAUGAUCAUUUGAUCAAUUUGAUCGAAUUGAACUUCUAAUGACCAGCGAGUAAAUAACUAAAGAAAUGAACACGUUAGUGAACUCUGUAGCUGAGUUUAUUCACUCACA ((-(((.((((((.....((((((((.....)))))))))))..))))))))....(.((((.((((..............(((....)))(((((.......))))))))).)))).). ( -23.30) >consensus AA_UUUCGAACCAUGAUCAUUUGAUCAGAUUGAUCGAAAUGAACUUCUAAUGACCAGCGAGUGAAUAACUGAAGAAAUGAACACGUUAGUGAACCGAGUAUGUGAGUUUAUUCACUCGCA ................((((((((((.....))))))).)))..............((((((((((((.............(((....)))................)))))))))))). (-16.05 = -18.29 + 2.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:01 2006