
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,433,462 – 9,433,553 |
| Length | 91 |
| Max. P | 0.877359 |

| Location | 9,433,462 – 9,433,553 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -13.81 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
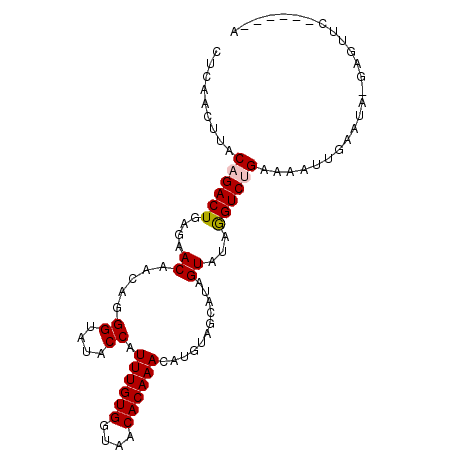
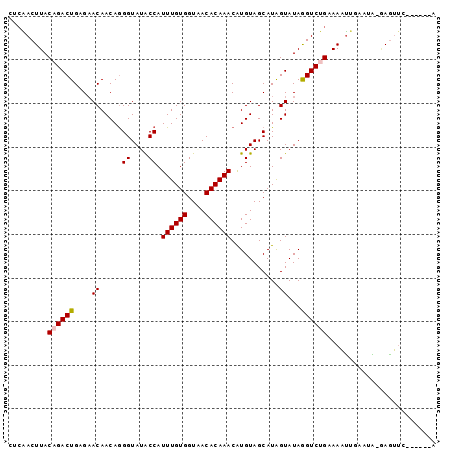
>2R_DroMel_CAF1 9433462 91 + 20766785 CUCGACUUACAGACUGAGAACAACAGGGUAUGCCAUUUGUGGUUACACAAACAUGUAGCAUAGUAUAGGUCUGAAAAUCGCAGG-AAAUUC------A (((((.((.((((((....((((...((....))..)))).((((((......))))))........)))))).)).))).)).-......------. ( -20.50) >DroVir_CAF1 28337 87 + 1 --CAACUUACAGACUGUGAACAACAGGGUAUACCAUUUGUGGUAACACAAACAUGUAGCAUGGUAUAGGUCUGUAAAUUUAAUA-UAGUU-------- --....(((((((((((.....))....(((((((((((((....))))).........)))))))))))))))))........-.....-------- ( -22.50) >DroGri_CAF1 28729 88 + 1 --AAACUUACAGACUGUGUACAACAAGGUAUACCAUUUGUGGUAACACAAACGUGUAGCAUGGUAUAAGUCUGUAAAUUCAUUAAGAGUU-------- --....(((((((((.(((((..((.(.(((((..((((((....)))))).))))).).))))))))))))))))((((.....)))).-------- ( -26.00) >DroYak_CAF1 20956 91 + 1 GUCGACUUACAGACUGAGAACAACAGGGUAUGCCAUUUGUGGUUACACAAACAUGUAGCAUAGUAUAGGUCUGAAAAUCGCAGG-AAGUUA------A (.(((.((.((((((....((((...((....))..)))).((((((......))))))........)))))).)).))))...-......------. ( -19.80) >DroMoj_CAF1 36906 97 + 1 CACAACUUACUGACUGUGAACAACAGGGUAUACCAUUUGUGGUAACACAAACAUGUAGCAUGGUAUAGGUCUGCAAAUUUAAUA-GAGUACUCCGUGA (((....((((..((((.....)))).(((.(((.((((((....))))))((((...)))).....))).)))..........-.))))....))). ( -21.00) >DroAna_CAF1 20686 91 + 1 CUCGACUUACUGACUGAGAACAACAGGGUAUGCCAUUUGUGGUGACACAAACAUGUAGCAUAGUAUAGGUCUGAAAUCGGUAUG-GAGAUC------A .((..(.((((((.(.(((.......(.(((((((((((((....)))))..)))..))))).).....))).)..)))))).)-..))..------. ( -22.80) >consensus CUCAACUUACAGACUGAGAACAACAGGGUAUACCAUUUGUGGUAACACAAACAUGUAGCAUAGUAUAGGUCUGAAAAUUGAAUA_GAGUUC______A .........((((((....((.....((....)).((((((....))))))...........))...))))))......................... (-13.81 = -14.50 + 0.69)

| Location | 9,433,462 – 9,433,553 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
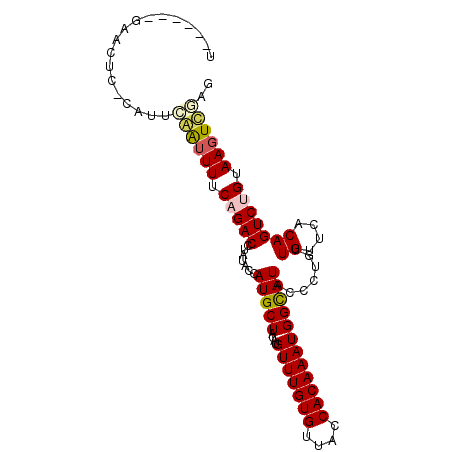
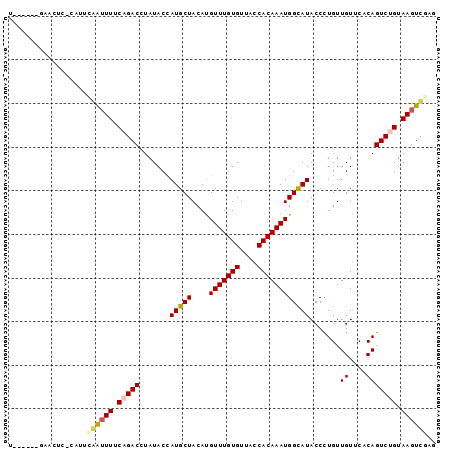
>2R_DroMel_CAF1 9433462 91 - 20766785 U------GAAUUU-CCUGCGAUUUUCAGACCUAUACUAUGCUACAUGUUUGUGUAACCACAAAUGGCAUACCCUGUUGUUCUCAGUCUGUAAGUCGAG .------......-....((((((.(((((......((((((....(((((((....)))))))))))))......((....))))))).)))))).. ( -22.10) >DroVir_CAF1 28337 87 - 1 --------AACUA-UAUUAAAUUUACAGACCUAUACCAUGCUACAUGUUUGUGUUACCACAAAUGGUAUACCCUGUUGUUCACAGUCUGUAAGUUG-- --------.....-.....(((((((((((.((((((((((.....))(((((....)))))))))))))...(((.....)))))))))))))).-- ( -22.50) >DroGri_CAF1 28729 88 - 1 --------AACUCUUAAUGAAUUUACAGACUUAUACCAUGCUACACGUUUGUGUUACCACAAAUGGUAUACCUUGUUGUACACAGUCUGUAAGUUU-- --------..........(((((((((((((((((.((...(((.((((((((....))))))))))).....)).))))...)))))))))))))-- ( -25.10) >DroYak_CAF1 20956 91 - 1 U------UAACUU-CCUGCGAUUUUCAGACCUAUACUAUGCUACAUGUUUGUGUAACCACAAAUGGCAUACCCUGUUGUUCUCAGUCUGUAAGUCGAC .------......-....((((((.(((((......((((((....(((((((....)))))))))))))......((....))))))).)))))).. ( -22.10) >DroMoj_CAF1 36906 97 - 1 UCACGGAGUACUC-UAUUAAAUUUGCAGACCUAUACCAUGCUACAUGUUUGUGUUACCACAAAUGGUAUACCCUGUUGUUCACAGUCAGUAAGUUGUG .(((((..((((.-..........((((...((((((((((.....))(((((....)))))))))))))..))))...........))))..))))) ( -21.45) >DroAna_CAF1 20686 91 - 1 U------GAUCUC-CAUACCGAUUUCAGACCUAUACUAUGCUACAUGUUUGUGUCACCACAAAUGGCAUACCCUGUUGUUCUCAGUCAGUAAGUCGAG .------......-.....((((((...........((((((....(((((((....)))))))))))))..((((((....))).))).)))))).. ( -17.20) >consensus U______GAACUC_CAUUCAAUUUUCAGACCUAUACCAUGCUACAUGUUUGUGUUACCACAAAUGGCAUACCCUGUUGUUCACAGUCUGUAAGUCGAG ..................((((((.(((((.......(((((....(((((((....)))))))))))).......((....))))))).)))))).. (-15.22 = -15.33 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:58 2006