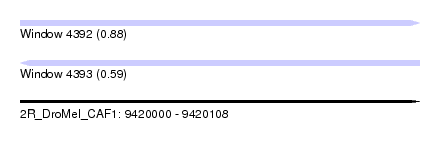
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,420,000 – 9,420,108 |
| Length | 108 |
| Max. P | 0.878635 |

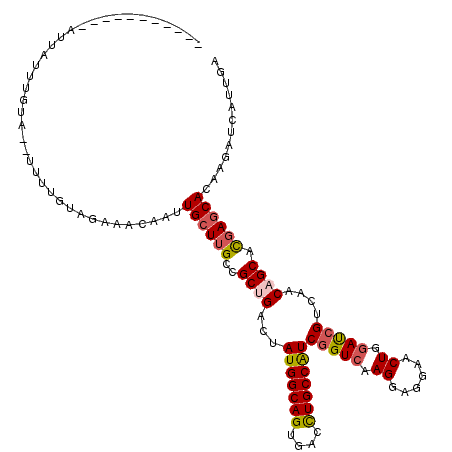
| Location | 9,420,000 – 9,420,108 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9420000 108 + 20766785 UCAAUGAUCUUGUGCUCGUGCUGUUGGCGAUCCAGUUCCUCCUUGACCGACGGCAGGUCACUGCCAUAGUCAGCGGCAAGCAAUUGUUUCUGCAACA--CGAAAACAAUC---------- .........((((..((((((((((((((.((.((......)).)).)...(((((....)))))...)))))))))..(((........)))...)--)))..))))..---------- ( -33.70) >DroVir_CAF1 12086 113 + 1 UCAAUGAUCUUGUGCUCGUGCUGUUGACGAUCCAGUUCCUCCUUGACCGAUGGCAGAUCACUGCCAUAGUCAGCGGCAAGCAAUUGUUUCUACAAAA--AAGAGGAAAUU-A---UAGA- .......((((.((((..(((((((((((.((.((......)).)).).(((((((....))))))).)))))))))))))).((((....))))..--)))).......-.---....- ( -33.40) >DroGri_CAF1 12794 107 + 1 UCGAUGAUCUUGUGCUCGUGCUGUUGACGAUCCAGUUCCUCCUUGACCGAUGGCAGAUCACUGCCAUAGUCGGCGGCAAGCAAUUGUUUCUGCAAAU--UACAAAUAAU----------- .........(((((((..(((((((((((.((.((......)).)).).(((((((....))))))).)))))))))))))..((((....))))..--.)))).....----------- ( -31.50) >DroWil_CAF1 9950 117 + 1 UCAAUAAUCUUGUGCUCAUGCUGUUGACGAUCCAGUUCAUCCUUGACCGAUGGCAAAUCACUGCCAUAGUCAGCGGCUAGCAAUUGUUUCUAUAAA---UGAUGACAAUCCAAAUUAGUU ....((((.(((((((...(((((((((......(.(((....))).).((((((......)))))).))))))))).))))((((((((......---.)).)))))).)))))))... ( -31.00) >DroMoj_CAF1 19536 112 + 1 UCAAUGAUUUUAUGUUCGUGCUGUUGACGAUCCAGUUCCUCCUUGACAGAUGGCAGGUCACUGCCAUAGUCAGCGGCAAGCAAUUGUUUCUAGAAUCACAACAAUAAG-------UAGU- ....(((((((((((...((((((((((......(((.......)))..(((((((....))))))).)))))))))).)))........))))))))..........-------....- ( -33.00) >DroAna_CAF1 9155 103 + 1 UCGAUGAUCUUGUGCUCGUGCUGUUGCCGGUCCAGUUCCUCCUUCACCGAUGGCAAGUCGCUGCCAUAGUCGGCGGCAAGCAAUUGUUUCUACAAUG-------AUAAUU---------- ......(((((((((...(((((((((((((..((......))..))).((((((......))))))....)))))).))))...))....)))).)-------))....---------- ( -29.10) >consensus UCAAUGAUCUUGUGCUCGUGCUGUUGACGAUCCAGUUCCUCCUUGACCGAUGGCAGAUCACUGCCAUAGUCAGCGGCAAGCAAUUGUUUCUACAAAA__AAAAAAAAAU___________ ............((((..((((((((((((......))...........(((((((....))))))).)))))))))))))).((((....))))......................... (-26.66 = -26.72 + 0.06)

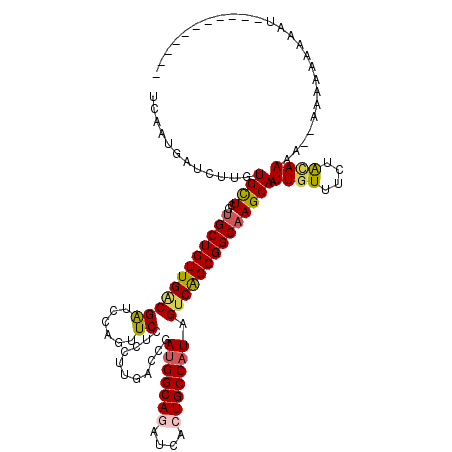
| Location | 9,420,000 – 9,420,108 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.07 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9420000 108 - 20766785 ----------GAUUGUUUUCG--UGUUGCAGAAACAAUUGCUUGCCGCUGACUAUGGCAGUGACCUGCCGUCGGUCAAGGAGGAACUGGAUCGCCAACAGCACGAGCACAAGAUCAUUGA ----------..((((.((((--(((((..((..((.((.(((.((..((((((((((((....))))))).))))).))))))).))..)).....))))))))).))))......... ( -36.90) >DroVir_CAF1 12086 113 - 1 -UCUA---U-AAUUUCCUCUU--UUUUGUAGAAACAAUUGCUUGCCGCUGACUAUGGCAGUGAUCUGCCAUCGGUCAAGGAGGAACUGGAUCGUCAACAGCACGAGCACAAGAUCAUUGA -((((---(-((.........--..)))))))......((((((..((((...(((((((....)))))))(((((.((......)).)))))....)))).))))))(((.....))). ( -33.60) >DroGri_CAF1 12794 107 - 1 -----------AUUAUUUGUA--AUUUGCAGAAACAAUUGCUUGCCGCCGACUAUGGCAGUGAUCUGCCAUCGGUCAAGGAGGAACUGGAUCGUCAACAGCACGAGCACAAGAUCAUCGA -----------....(((((.--....)))))......((((((((.(((((((((((((....))))))).))))..)).))..(((.........)))..))))))............ ( -29.00) >DroWil_CAF1 9950 117 - 1 AACUAAUUUGGAUUGUCAUCA---UUUAUAGAAACAAUUGCUAGCCGCUGACUAUGGCAGUGAUUUGCCAUCGGUCAAGGAUGAACUGGAUCGUCAACAGCAUGAGCACAAGAUUAUUGA ......((((((((((..((.---......)).))))))(((....((((((((((((((....))))))).))))...(((((......)))))...)))...))).))))........ ( -28.20) >DroMoj_CAF1 19536 112 - 1 -ACUA-------CUUAUUGUUGUGAUUCUAGAAACAAUUGCUUGCCGCUGACUAUGGCAGUGACCUGCCAUCUGUCAAGGAGGAACUGGAUCGUCAACAGCACGAACAUAAAAUCAUUGA -....-------....((((((((((.((((...(((....)))((..((((.(((((((....)))))))..)))).)).....)))))))).)))))).................... ( -28.50) >DroAna_CAF1 9155 103 - 1 ----------AAUUAU-------CAUUGUAGAAACAAUUGCUUGCCGCCGACUAUGGCAGCGACUUGCCAUCGGUGAAGGAGGAACUGGACCGGCAACAGCACGAGCACAAGAUCAUCGA ----------....((-------((((((....)))))((((((..(((......))).((...(((((...(((..((......))..))))))))..)).))))))...)))...... ( -26.30) >consensus ___________AUUAUUUGUA__UUUUGUAGAAACAAUUGCUUGCCGCUGACUAUGGCAGUGACCUGCCAUCGGUCAAGGAGGAACUGGAUCGUCAACAGCACGAGCACAAGAUCAUUGA ......................................((((((..((((...(((((((....)))))))(((((.((......)).)))))....)))).))))))............ (-24.70 = -25.07 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:54 2006