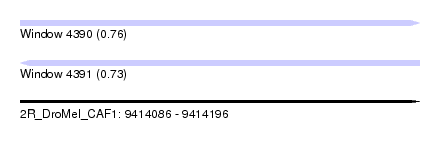
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,414,086 – 9,414,196 |
| Length | 110 |
| Max. P | 0.764472 |

| Location | 9,414,086 – 9,414,196 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.66 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764472 |
| Prediction | RNA |
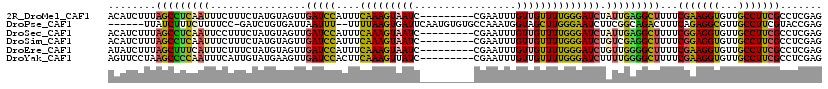
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9414086 110 + 20766785 CUCGAGGCGAAGGCAACACCUUCGAAAAGCCUCAAUAGAUCCCAAAACAACAAAUUCG---------GAUUACUUUGAAAUGGAUCAACUACAUAGAAAGAAAUUGAGGCUAAAGAUGU .......((((((.....))))))...(((((((((.(((((............((((---------((....))))))..)))))..((....))......)))))))))........ ( -26.04) >DroPse_CAF1 3891 110 + 1 CUCGGUACGAAGGCAACGCCUCUGAAAGUCUGCCGAAGAUUCCCAAGCUACCAUUUGGCACACAUUGAAUCACUUAAAA--AAAUUAAUCACAGAUC-GGAAAAGAAAGAUAA------ .((....(((.(((...)))((((...((.(((((((................))))))).)).((((.....))))..--..........))))))-).....)).......------ ( -16.69) >DroSec_CAF1 3338 110 + 1 CUCGAGGCGAAGGCAACACCUCCGAAAAGCCUCAAUAGAUCCCAAAACAACAAAUUCG---------GAUUACUUUGAAAUGGAUCAACUACAUAGAAAGGAAUUGAGGCUAAAGAUGU .((((((....(....).))).)))..(((((((((.(((((...............)---------)))).((((...(((.........)))...)))).)))))))))........ ( -23.66) >DroSim_CAF1 10011 110 + 1 CUCGAGGCGAAGGCAACACCUCCGAAAAGCCUCGACAGAUCCCAAAACAACAAAUUCG---------GAUUACUUUGAAAUGGAUCAACUACAUAGAAAGAAAUUGAGGCUAAAGAUGU .((((((....(....).))).)))..((((((((..(((((............((((---------((....))))))..)))))..((....)).......))))))))........ ( -21.74) >DroEre_CAF1 3255 110 + 1 CUCGAGGCGAAGGCAACACCUUCGAAAAGCCCCAACAGAUCCCAAAACAACAAAUUCG---------GAUUACUUUGAAAUGGAUCAACUACAUAGAAAGAAAUGAAAGCUAAAGAUAU .......((((((.....))))))...(((.......(((((............((((---------((....))))))..))))).....(((........)))...)))........ ( -15.74) >DroYak_CAF1 3361 110 + 1 CUCGAGGCGAAGGCAACACCUUCGAAAAGCCCCAAAAGAUCCCAAAACAACAAAUUCG---------GAUAACUUUGAAGUGGAUCAACUUCAUACAAUGAAAUUGGGGCUUAGGAACU .......((((((.....))))))..(((((((((..(((((............((((---------((....))))))..)))))...(((((...))))).)))))))))....... ( -31.24) >consensus CUCGAGGCGAAGGCAACACCUCCGAAAAGCCUCAAAAGAUCCCAAAACAACAAAUUCG_________GAUUACUUUGAAAUGGAUCAACUACAUAGAAAGAAAUUGAGGCUAAAGAUGU .((((((....(....)..))))))..((((((((..(((((.......................................)))))..((........))...))))))))........ (-14.69 = -14.66 + -0.02)

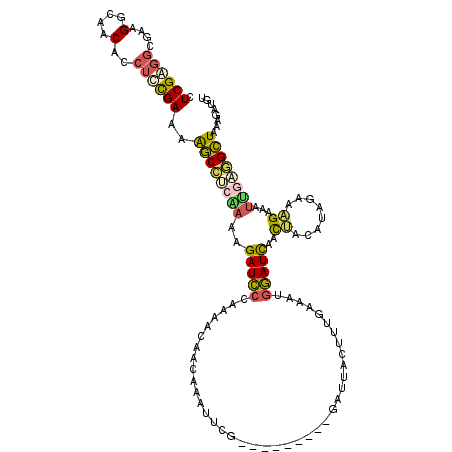
| Location | 9,414,086 – 9,414,196 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -20.34 |
| Energy contribution | -19.41 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9414086 110 - 20766785 ACAUCUUUAGCCUCAAUUUCUUUCUAUGUAGUUGAUCCAUUUCAAAGUAAUC---------CGAAUUUGUUGUUUUGGGAUCUAUUGAGGCUUUUCGAAGGUGUUGCCUUCGCCUCGAG ........(((((((((.((....(((....((((......)))).)))..(---------((((........)))))))...)))))))))..((((.((((.......)))))))). ( -27.80) >DroPse_CAF1 3891 110 - 1 ------UUAUCUUUCUUUUCC-GAUCUGUGAUUAAUUU--UUUUAAGUGAUUCAAUGUGUGCCAAAUGGUAGCUUGGGAAUCUUCGGCAGACUUUCAGAGGCGUUGCCUUCGUACCGAG ------..............(-(.(((((..((((...--..))))..(((((...((.((((....))))))....)))))....)))))......(((((...))))).....)).. ( -18.90) >DroSec_CAF1 3338 110 - 1 ACAUCUUUAGCCUCAAUUCCUUUCUAUGUAGUUGAUCCAUUUCAAAGUAAUC---------CGAAUUUGUUGUUUUGGGAUCUAUUGAGGCUUUUCGGAGGUGUUGCCUUCGCCUCGAG ........(((((((((.......(((....((((......)))).))).((---------((((........))))))....))))))))).....((((((.......))))))... ( -27.60) >DroSim_CAF1 10011 110 - 1 ACAUCUUUAGCCUCAAUUUCUUUCUAUGUAGUUGAUCCAUUUCAAAGUAAUC---------CGAAUUUGUUGUUUUGGGAUCUGUCGAGGCUUUUCGGAGGUGUUGCCUUCGCCUCGAG ..(((((.(((..(((.(((....(((....((((......)))).)))...---------.))).)))..)))..)))))...(((((((.....(((((.....)))))))))))). ( -26.90) >DroEre_CAF1 3255 110 - 1 AUAUCUUUAGCUUUCAUUUCUUUCUAUGUAGUUGAUCCAUUUCAAAGUAAUC---------CGAAUUUGUUGUUUUGGGAUCUGUUGGGGCUUUUCGAAGGUGUUGCCUUCGCCUCGAG ........((((..(((........))).))))(((((....(((((((((.---------.......))))))))))))))..(((((((.....(((((.....)))))))))))). ( -24.50) >DroYak_CAF1 3361 110 - 1 AGUUCCUAAGCCCCAAUUUCAUUGUAUGAAGUUGAUCCACUUCAAAGUUAUC---------CGAAUUUGUUGUUUUGGGAUCUUUUGGGGCUUUUCGAAGGUGUUGCCUUCGCCUCGAG .......(((((((((..((......((((((......)))))).......(---------((((........)))))))....))))))))).((((.((((.......)))))))). ( -33.20) >consensus ACAUCUUUAGCCUCAAUUUCUUUCUAUGUAGUUGAUCCAUUUCAAAGUAAUC_________CGAAUUUGUUGUUUUGGGAUCUAUUGAGGCUUUUCGAAGGUGUUGCCUUCGCCUCGAG ........(((((((((................(((((....(((((((((.................)))))))))))))).)))))))))...(((((((...)))))))....... (-20.34 = -19.41 + -0.93)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:52 2006