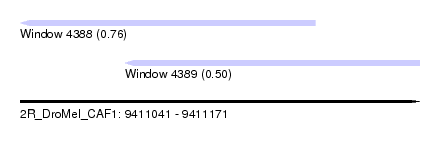
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,411,041 – 9,411,171 |
| Length | 130 |
| Max. P | 0.759728 |

| Location | 9,411,041 – 9,411,137 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.75 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
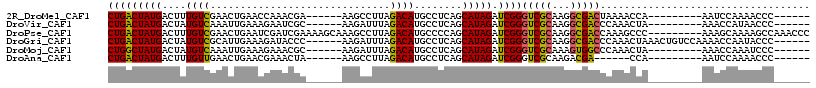
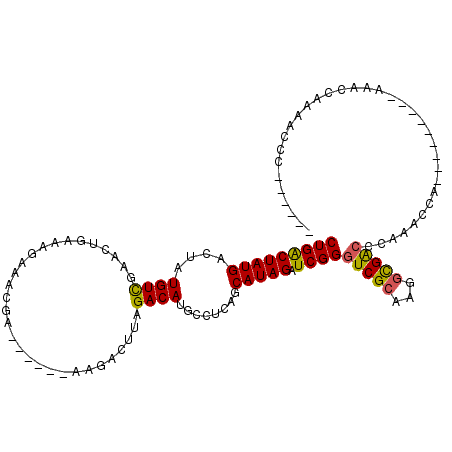
>2R_DroMel_CAF1 9411041 96 - 20766785 AGCCUUAGACAUGCCUCAGCAU---AGAUCGGGUCGCAAGGCGACUAAAACCA---------AAUCCAAAACCCAAACCACGACUGUCCAAGUGGCAGAGGAAAGUGA .........(((.((((.....---......((((((...)))))).......---------...............((((..........))))..))))...))). ( -20.60) >DroVir_CAF1 285 96 - 1 AGAUUUAGACAUGCCUCAGCAU---AGAUCGGGUCGCAAGGCGACCCAAACUA---------AAACCAUAACCCAAACCACAACUGCCUAUGUGGCAGAGGAAAGUGA .........(((.((((.....---.....(((((((...)))))))......---------......................((((.....))))))))...))). ( -27.90) >DroGri_CAF1 288 105 - 1 AGAUUUAGACAUGCCUCAGCAU---AGAUCGGGUCGCAAGGCGACCCAAACUAAACUGUCCAAAACCAAUACCCAAACCACAACUGUCCAAGUGGCAGAGGAAAGUGA .........(((.((((((..(---((...(((((((...)))))))...)))..))....................((((..........))))..))))...))). ( -25.90) >DroWil_CAF1 314 93 - 1 AGCCUUAGCCAUGCCUCAGCAUACUAAAACGAGUCGUAAGACGACUAAAACC---------------AAAACCCAAACAACAACUGUCCAAGUGGCAGAGGAAAGUGA ..((((.(((((((....))...........(((((.....)))))......---------------........................))))).))))....... ( -19.50) >DroMoj_CAF1 6954 96 - 1 AGAUUUAGACAUGCCUCAGCAU---AGAUCGGGUCGCAAAGUGGCCCAAACUA---------AAACCAAAUCCCAUACCACAACUGUCCAUGUGGCAGAGGAAAGUGA ...(((((..((((....))))---.....(((((((...)))))))...)))---------))..((..(((..(.(((((........))))).)..)))...)). ( -25.30) >DroAna_CAF1 312 90 - 1 AGCCUUAGACAUGCCUCAGCAU---AGAUCGGGUCGCAAGACGA------CCA---------AAUCCAAAACCCAAACCACGACUGUCCAAGUGGCAGAGGAAAGUGA .........(((.((((.....---.(((..(((((.....)))------)).---------.)))...........((((..........))))..))))...))). ( -22.90) >consensus AGACUUAGACAUGCCUCAGCAU___AGAUCGGGUCGCAAGGCGACCCAAACCA_________AAACCAAAACCCAAACCACAACUGUCCAAGUGGCAGAGGAAAGUGA .........(((.((((..............((((((...))))))...............................((((..........))))..))))...))). (-17.86 = -17.75 + -0.11)


| Location | 9,411,075 – 9,411,171 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9411075 96 - 20766785 CUGACUAUGACUUUGUCGAACUGAACCAAACGA------AAGCCUUAGACAUGCCUCAGCAUAGAUCGGGUCGCAAGGCGACUAAAACCA---------AAUCCAAAACCC------ (((((((((....(((((........)...(..------..).....))))........))))).))))(((((...)))))........---------............------ ( -16.90) >DroVir_CAF1 319 96 - 1 CUGACUAUGACUAUGUCAAAUUGAAAGAAUCGC------AAGAUUUAGACAUGCCUCAGCAUAGAUCGGGUCGCAAGGCGACCCAAACUA---------AAACCAUAACCC------ ..(((((((..((((((...((....))(((..------..)))...))))))......))))).))(((((((...)))))))......---------............------ ( -26.70) >DroPse_CAF1 332 108 - 1 CUGACUAUGACUUUGUCGAACUGAAUCGAUCGAAAAGCAAAGCCUUAGACAUGCCCCAGCAUAGAUCGGGUCGCAAGGCGACCAAAGCCC---------AAAGCAAAAGCCAAACCC .......((.(((((...........(((((...(((......)))....((((....)))).)))))((((((...))))))......)---------))))))............ ( -22.60) >DroGri_CAF1 322 105 - 1 CUGACUAUGACUAUGUCGCAUUGAAAGAUACCC------AAGAUUUAGACAUGCCUCAGCAUAGAUCGGGUCGCAAGGCGACCCAAACUAAACUGUCCAAAACCAAUACCC------ ..(((...((((((((.((((....((((....------...))))....))))....)))))).))(((((((...)))))))..........)))..............------ ( -23.50) >DroMoj_CAF1 6988 96 - 1 CUGGCUAUGACUAUGUCAAAUUGAAAGAAACGC------AAGAUUUAGACAUGCCUCAGCAUAGAUCGGGUCGCAAAGUGGCCCAAACUA---------AAACCAAAUCCC------ .((((((((..((((((...((....))..(..------..).....))))))......)))))...(((((((...)))))))......---------...)))......------ ( -23.30) >DroAna_CAF1 346 90 - 1 CUGACUAUGACUUUGUUGAACUGAACGAAACUA------AAGCCUUAGACAUGCCUCAGCAUAGAUCGGGUCGCAAGACGA------CCA---------AAUCCAAAACCC------ (((((((((..((((((......)))))).(((------(....))))...........))))).))))(((....)))..------...---------............------ ( -18.00) >consensus CUGACUAUGACUAUGUCGAACUGAAAGAAACGA______AAGACUUAGACAUGCCUCAGCAUAGAUCGGGUCGCAAGGCGACCCAAACCA_________AAACCAAAACCC______ (((((((((....((((..............................))))........))))).))))(((((...)))))................................... (-12.85 = -12.63 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:50 2006