| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,660,543 – 1,660,683 |
| Length | 140 |
| Max. P | 0.981982 |

| Location | 1,660,543 – 1,660,663 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -37.10 |
| Energy contribution | -37.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1660543 120 + 20766785 CUAGAUAUGAAAUAUCGAACUGGUUCCAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGUAUUUAUUAUUUUUC ..(((((..((((((((((....)))....(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))))))))))..)))))... ( -43.60) >DroSec_CAF1 46154 113 + 1 GUA-------AAUAUCAAUCUGGUUUCAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGUGAAUUAUUAUUUUUA ...-------..............(((((.(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))))))))............ ( -39.90) >DroEre_CAF1 50290 116 + 1 GUAAAUAUAAAAUGUCAUGCAGGUUUCAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGC--UUUUUAUUUU-- ......((((((.((((((.......))).(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))))))--.))))))...-- ( -40.60) >consensus GUA_AUAU_AAAUAUCAAACUGGUUUCAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGCA_UUAUUAUUUUU_ ..............................(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))................. (-37.10 = -37.10 + 0.00)


| Location | 1,660,543 – 1,660,663 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1660543 120 - 20766785 GAAAAAUAAUAAAUACCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUGGAACCAGUUCGAUAUUUCAUAUCUAG ((((.............(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))(((.(((....))).)))))))........ ( -35.50) >DroSec_CAF1 46154 113 - 1 UAAAAAUAAUAAUUCACGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUGAAACCAGAUUGAUAUU-------UAC ............((((.(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))..))))..............-------... ( -32.00) >DroEre_CAF1 50290 116 - 1 --AAAAUAAAAA--GCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUGAAACCUGCAUGACAUUUUAUAUUUAC --...((((((.--...(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))((((.......))))...))))))...... ( -32.40) >consensus _AAAAAUAAUAA_UACCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUGAAACCAGAUUGAUAUUU_AUAU_UAC .................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).............................. (-30.80 = -30.80 + 0.00)

| Location | 1,660,583 – 1,660,683 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1660583 100 + 20766785 CGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGUAUUUAUUAUUUUUC-AUGUUCUUUCAUUAUUUAUA (((..((((...(.(..((((((..((((((....))))))..).)))))..).)))))..)))................-.................... ( -25.40) >DroSec_CAF1 46187 97 + 1 CGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGUGAAUUAUUAUUUUUAGACGUUCUUU----UUUUAUA ......(((((.((((.(((((....(((((....)))))((((.......))))))))).)))).)))))...................----....... ( -27.00) >DroEre_CAF1 50330 87 + 1 CGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGC--UUUUUAUUUU--------GUUU----AUAUGUA .((((.(((..(((....(((((..((((((....))))))..).))))((((......)))).))--)..)))..))--------))..----....... ( -27.30) >consensus CGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGCA_UUAUUAUUUUU__A_GUUCUUU____AUUUAUA (((..((((...(.(..((((((..((((((....))))))..).)))))..).)))))..)))..................................... (-25.17 = -25.17 + 0.00)

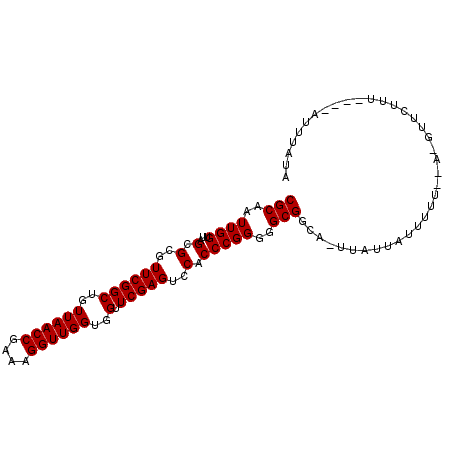
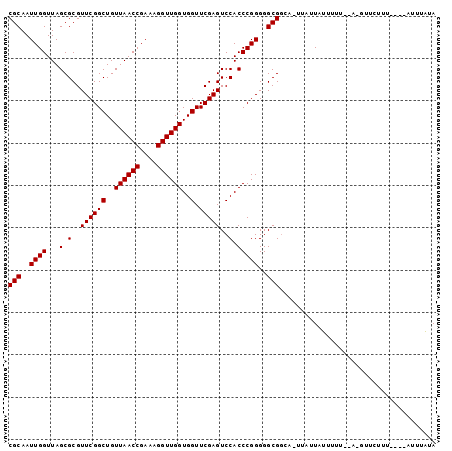
| Location | 1,660,583 – 1,660,683 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1660583 100 - 20766785 UAUAAAUAAUGAAAGAACAU-GAAAAAUAAUAAAUACCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCG ..........(((((.....-...............(((((....)))))....(......)...)))))(((((...((.....))..)))))....... ( -18.80) >DroSec_CAF1 46187 97 - 1 UAUAAAA----AAAGAACGUCUAAAAAUAAUAAUUCACGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCG .......----..........................(((.....(((((.......))))).....((((((.....)))))).)))............. ( -17.70) >DroEre_CAF1 50330 87 - 1 UACAUAU----AAAC--------AAAAUAAAAA--GCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCG .......----....--------.........(--((.((.....(((((.......))))).....((((((.....)))))).)))))........... ( -19.00) >consensus UAUAAAU____AAAGAAC_U__AAAAAUAAUAA_UACCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCG .....................................(((.....(((((.......))))).....((((((.....)))))).)))............. (-17.57 = -17.57 + -0.00)

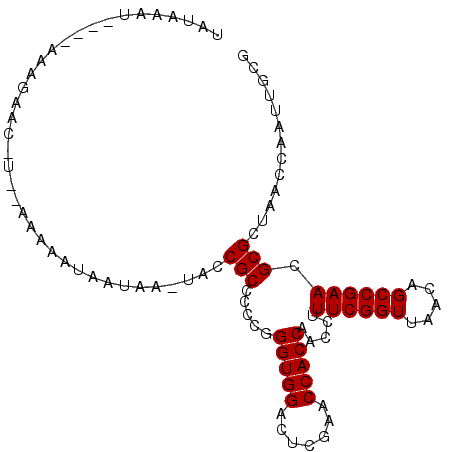
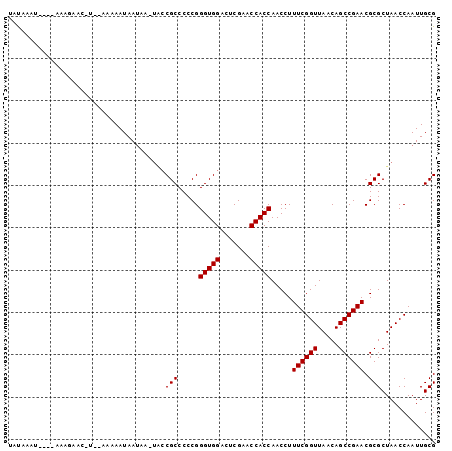
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:05 2006